XLUM-file Format Documentation
1 Motivation and principles
This document details the XLUM format, an XML based file format for long-term data preservation and exchange of luminescence data. The format is readable by humans and machines, and the data can be easily checked with any text reader on any major operating system. This design allows a platform-independent operation. The following documentation provides essential information on the file XLUM format and accounts for the needs for individual flexible data analysis, even with self-written software applications.
Three simple design principles underpin the format specification:
- Stored are data recorded over time or time derived instances,
- stored are data on a technical component level,
- data stored in a file are self-consistent.
2 General description
The XLUM-format is an XML derivative. The base data structure is a
tree with five nodes types storing the data. Each level has its unique denotation.
The levels and a short introduction is given in the table below. A detailed description can be found in the following sections.
Nodes represent each level in the structure. Only <curve> nodes contain physical quantities, such as luminescence data all other nodes are parent nodes of <curve>
to structure the dataset and ship additional metadata.
The first level, xlum is a root node to enable storage of luminescence data in arbitrary files following the XML scheme. As a side effect this also allows custom file endings different from *.xlum.
| Node name | Number of samples | Number of aliquots | Number of records | Number of data curves |
|---|---|---|---|---|
<xlum/> | \(\inf\) | \(\inf\) | \(\inf\) | \(\inf\) |
<sample/> | 1 | \(\inf\) | \(\inf\) | \(\inf\) |
<sequence/> | 1 | 1 | \(\inf\) | \(\inf\) |
<record/> | 1 | 1 | 1 | \(\inf\) |
<curve/> | 1 | 1 | 1 | 1 |
A node has a name, attributes, and data stored in it. The data stored in the attributes describe the state of this level. The stored data in the node describes the process(es) assigned to the node. A minimal example is shown in the listing. The documentation provides an overview starting with the leaf node description and going up to the root from there. Further notes:
- The format version bases on XML version 1.0 (Fifth Edition)
- File encoding should always be UTF-8
- This specification lists only mandatory attributes. Additional, custom attributes are explicitly supported.
- Parser supporting the
XLUMformat must not crash when encountering non-specified node attributes. However, they may skip them.
<?xml version="1.0" encoding="utf-8"?>
<xlum>
<sample>
<sequence>
<record>
<curve>
1 2 ... n
</curve>
</record>
</sequence>
</sample>
</xlum>3 Detailed description
3.1 The <curve/> level
The <curve/> level is the deepest node (leaf) and has no further sub-levels.
A curve holds the predefined/simulated or measured output of one single
technical component. For example, a typical thermoluminescence measurement
may consist of one or many curves.
The one curve example: Time against temperature. In this case temperature is a processed quantity, because measurements happen over a time instant. However, for compatibility reasons, this would be allowed although it is not preferred.
The three curve example: (a) Time against temperature recorded by a thermocouple (temperature sensor), (b) time against photon counts recorded by a photomultiplier tube, (c) time against a predefined heating ramp.
In both cases, all mentioned <curve/> nodes belong to one parent <record/>. In case 1,
the record contains one curve and three in case 2. Ideally, curves represent technical
components and not processed quantities.
3.1.1 Value storage
Numerical values in the <curve/> node are stored white space separated. Only quantities detected
by a detector/sensor are stored in this node. This includes simulated values.
Figure 3.1 shows three types of (luminescence) detectors, distinguished by their capacity to record spatially resolved information.
Regardless of the detector, recorded quantities are stored in the <curve/> node
separated by white space. Example:
<curve>59.5167 109.5164 149.7003 109.5170 156.2654</curve>Values in the node are real numbers (-1e-307 \(\leqslant x \leqslant\) 1e+307; \(x \in \mathbb{R}\)).
Scientific ‘E’/‘e’ notations of numbers are explicitly allowed, example: 1e+2 for 100 or 1e-2 for 0.01.
Decimal separator is a . (dot). Other separators, for instance to separate groups of numbers
are not allowed to avoid decimal delimiter problems. Examples:
- 10000.00 -> OK
- 10 000.00 -> NOT OK
- 10,000.00 -> NOT OK
To reduce the size of the file, for instance for image data, the string in <curve/> can be stored using base64 encoding. It is up to the parser to decide whether the string
was encoded or not! Encoding in other places (attributes) is not supported and must not be expected by
the parser.
Please note that the XSD document does not specify binary encoding because it considers all values in <curve/> strings (which can represent encoded values).
For consistency reasons, the value in the node span an array with dimensions
defined through the node attributes xValues, yValues and tValues (see Fig. 3.1).
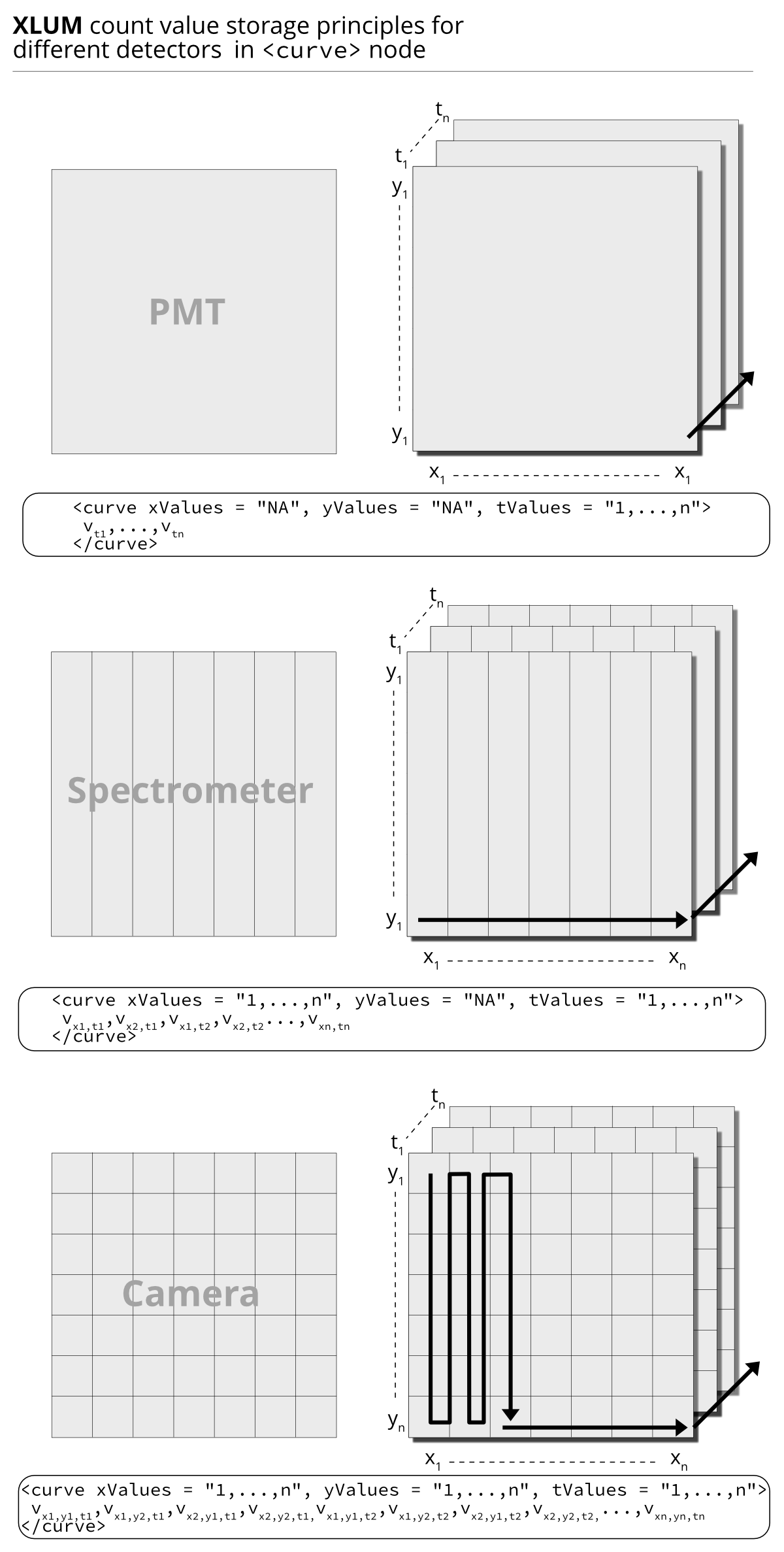
Figure 3.1: The three cases for data storage depending on the detector. The black solid arrows indicate how the array is constructed from the values stored in the node.
The simplest form is a detector that records no spatial information. For instance, a photomultiplier tube only knows about count recorded over time. Alternatively, the detector can be any sensor, measuring, e.g., temperature, pressure or current.
A spectrometer (here a camera in front of a spectrograph) is another type of detector with spatial information, here wavelengths split by the spectrograph hitting different pixel areas of the detector.
The most complex from is a detector that records events (e.g., luminescence), spatially resolved. Such as a camera.
In the examples and Fig.3.1 the detectors detect luminescence. However, any type of sensor, electronic component recording information of relevance for the measurement of luminescence are suitable. Typical examples:
- Temperature sensor (thermocouple), recording the temperature of the heating element
- Photo diode, monitoring the optical power density at the sample position
- Resistor, measuring the current of a LED
- Pressure sensor, monitoring the atmosphere in the measurement chamber
- Inductive sensor, monitoring closing and opening of a shutter
3.1.2 Node attributes
| Identifier | Type | Allows NA? | Example | Information |
|---|---|---|---|---|
| component | string | yes | "EMI 9235QB15" | name of the technical component. NA allowed only for "predefined" |
| startDate | dateTime | no | "2021-07-14T22:59:35.0Z" | ISO 8601-1:2019: YYYY-MM-DDThh:mm:ss[.mmm]Z recalculated to Zulu time |
| curveType | string | no | "predefined" | Values allowed are only "predefined" or "measured" |
| duration | number | no | "20.000" | Duration of the detection in seconds or a fraction of it |
| offset | number | no | "10.000" | Before the detection starts in seconds or a fraction of it |
| xValues | list unsignedInt | no | "1 2 3" | x-coordinate values of the detector; 0 indicates that that this parameter is not used |
| yValues | list unsignedInt | no | "1 2 3" | y-coordinate values of detector; 0 indicates that that this parameter is not used |
| tValues | list double | no | "1. 2.0 3.0" | time values in seconds or fractions of it. Values must be positive |
| vLabel | string | no | "Luminescence" | Measured physical quantity |
| xLabel | string | yes | "pixel" | label of the x-coordinate values |
| yLabel | string | yes | "pixel" | label of the y-coordinate values |
| tLabel | string | no | "time" | label of t-values, usually ‘time’ |
| vUnit | string | no | "cts" | label of the v-values, for luminescence usually photon counts |
| xUnit | string | yes | "nm" | SI unit or equivalent for x-values |
| yUnit | string | yes | "px" | SI unit or equivalent for y-values |
| tUnit | string | no | "s" | SI unit or equivalent for t-values |
| detectionWindow | string | yes | "375" | Centre wavelength if applicable. can be set to NA |
| filter | list (string) | yes | "Hoya U340; Delta BP 365/50EX" | Filter names separated by ; |
3.1.3 Example
In order to define a suitable standard only a few attributes are required. A few of them are useful for every kind of component, others are only meaningful in combination. For instance, providing information on the detection window is not meaningful for a heating element.
{...}
<curve component="heating element" startDate="2021-02-14T22:57:00.0Z"
curveType="predefined" duaration="5"
offset="0" xValues="NA" yValues="NA"
tValues="1 2 3 4 5" xLabel="NA" yLabel="NA"
tLabel="time" vLabel="temperature" xUnit="NA" yUnit="NA" vUnit="K" tUnit="s">
293 303 313 323 333
</curve>
<curve component="thermo couple" startDate="2021-02-14T22:57:00.0Z"
curveType="measured" duaration="5"
offset="0" xValues="NA" yValues="NA" tValues="1 2 3 4 5" xLabel="NA" yLabel="NA"
vLabel="temperature" tLabel="time" xUnit="NA" yUnit="NA" vUnit="K" tUnit="s">
293 303 313 323 333
</curve>
<curve component="EMI 9123QB15" startDate="2021-02-14T22:57:00.0Z"
curveType="measured" duaration="5" offset="0" xValues="NA" yValues="NA"
tValues="1 2 3 4 5" xLabel="NA" yLabel="NA"
vLabel="luminescence" tLabel="time" xUnit="NA" yUnit="NA" vUnit="cts"
tUnit="s" detectionWindow="380 nm" filter="Hoya U340 + Delta 365/40">
20 25 32 41 46
</curve>
{...}One curve is related to one technical device. To define a accurate time window,
the parameters duration and offset should be used. These parameters are related to the
start of the parent <record/> node.
3.2 The <record/> level
A <record> defines one process involving many components, hence <record> may
contain \(1\leq n\leq Inf; n \in \mathbb{Z}\) <curve> nodes. A record can also be understood
as one step in a measurement sequence, e.g., a TL measurement. All curves within the record should have been
detected within the same time frame defined through the attributes <startDate> and the duration of the step.
3.2.1 Attributes
| Identifier | Type | Allows NA? | Example | Information |
|---|---|---|---|---|
| recordType | string | no | "TL" | valid values according to the recordType table |
| sequenceStepNumber | unsignedInt | yes | 5 | index in the sequence |
| sampleCondition | string | yes | dose | valid values according to the table below |
The recordType names are not carved in stone, but should be chosen to best describe a record in one
word or using an accepted abbreviation (e.g., OSL for optically stimulated luminescence).
However, it is important to keep in mind that the recordType makes no claim on the technical
components involved, but is solely set to ease the understanding of the record. The hard
physical information is stored in the <curve> node.
recordType | Information |
|---|---|
"bleaching" | any kind of bleaching |
"irradiation" | irradiation with ionising source |
"atmosphereExchange" | atmosphere exchange |
"heating" | heating without signal detection |
"spectrometer" | spectrometer |
"camera" | camera, e.g. any kind of imaging other than spectrometry |
"TL" | thermoluminescence measurement |
"ITL" | isothermal luminescence |
"IRSL" | infrared stimulated luminescence |
"TM-OSL" | thermally modulated optically stimmulated luminescence |
"RF" | radiofluorescence |
"UV-RF" | radiofluorescence with detection in the UV wavelength range |
"IR-RF" | radiofluorescence with detection in the infrared wavelength range |
"IR-PL" | infrared photoluminescence |
"OSL" | optically stimulated luminescence (wavelength unspecified) |
"BSL" | blue-light optically stimulated luminescence |
"GSL" | green-light optically stimulated luminescence |
"VSL" | violet-light optically stimulated luminescence |
"YSL" | yellow-light optically stimulated luminescence |
"POSL" | pulsed optical stimulated luminescence |
"pause" | pause |
"custom" | any undefined or custom record type not listed above |
Valid entries for for sampleConditions are given in the following table:
sampleCondition | Information |
|---|---|
"Natural" | natural luminescence signal |
"Natural+Dose" | natural luminescence signal with additive dose |
"Bleach" | artificially depleted luminescence |
"Bleach+Dose" | artificially depleted luminescence with an additive dose |
"Nat.(Bleach)" | artificially bleached natural luminescence signal |
"Nat.+Dose(Bleach)" | artificially bleached natural luminescence with added dose |
"Dose" | artificially irradiated sample |
"Background" | background |
Please note that the attribute sampleCondition is of informative nature, accounting
for information stored in BIN/BINX-files. The field can be set to NA.
3.2.2 Example
The following examples consists of two records.
{...}
<record recordType="GSL" sequenceStepNumber="1" sampleCondition="NA">
<curve ...>
{...}
</curve>
<curve ...>
{...}
</curve>
</record>
<record recordType="bleaching" sequenceStepNumber="2" sampleCondition="NA">
<curve ...>
{...}
</curve>
</record>
{...}3.3 The <sequence/> level
A sequence describes multiple measurements sequentially used at one aliquot,
for instance as cup/disc with multiple grains or a single grain. Hence, another
aliquot from the same sample and the sample sequence needs to be wrapped in a new <sequence/> node.
Naturally the information stored an that level can be very verbose, but are limited to
a few required attributes only. In the luminescence context a sequence can fit, e.g.,
a SAR protocol, but also a succession of protocols measured on one aliquot.
3.3.1 Attributes
| Identifier | Type | Allows NA? | Example | Information |
|---|---|---|---|---|
| position | numeric | no | "42" | position of aliquot in the reader, can be 0 in case samples are changed manually, i.e. the equipment has only one single measurement position |
| name | string | yes | "SAR measurement" | name of the sequence |
| fileName | string | yes | "SAR_OSL.seq" | sequence file so far applicable |
| software | string | yes | "Sequence Editor 4.4" | software used to write the sequence if applicable |
| readerName | string | yes | "Risø OSL/TL" | name of the luminescence reader employed for the measurement |
| readerSN | string | yes | "234-23" | reader serial number |
| readerFW | string | yes | "reader OSL 12.3" | reader firmware version |
3.3.2 Example
{...}
<sequence position="1" name="SAR measurment"
fileName="Testsequence.seq" software="DeviceStudio, 2.37">
<record ...>
<curve ...>
{...}
</curve>
{...}
</record>
{...}
</sequence>
{...}3.4 The <sample/> level
The sample constitutes the top level for one particular sample. The sample level may contain an infinite number of sequences measured for one sample.
3.4.1 Attributes
| Identifier | Data type | Allows NA? | Example | Information |
|---|---|---|---|---|
| name | number | yes | "LUM-21321" | unique sample identifier |
| mineral | string | yes | "quartz" | dominant mineral composition of the sample |
| latitude | numeric | yes | "52.4091392" | latitude in decimal degrees of the sample origin |
| longitude | numeric | yes | "-4.0702446" | longitude in decimal degrees of the sample origin |
| altitude | numeric | yes | "50" | altitude in m above see level |
| doi | anyURI | yes | "10.1016/j.radmeas.2017.02.003" | valid DOI with further information on the sample, if applicable |
3.4.2 Example
{...}
<sample name="LUM-21321" mineral="quartz" latitude="52.4091392" longitude="-4.0702446"
altitude="50" doi="10.1016/j.radmeas.2017.02.003">
<sequence ...>
<record ...>
<curve ...>
{...}
</curve>
{...}
</record>
{...}
</sequence>
{...}
</sample>
{...}3.5 The <xlum/> level
The <xlum> is the format parent node and it can contain an infinite number
of samples. Only this level allows to set a license for the distribution
and usage of the data. All data within a particular <xlum> are considered
distributed under the license. You have to create new instances of the <xlum>
to set different licenses.
3.5.1 Attributes
| Identifier | Type | Allows NA? | Example | Information |
|---|---|---|---|---|
| xmlns | namespace | no | predefined | Format specific namespace |
| lang | string | no | predefined | ISO 639-1 language code, must not be changed |
| version | numeric | no | "1.0" | the xlum format version number relevant |
| flavour | string | no | "generic" | allows to specify a particular flavour of the format, for instance for particular equipment |
| author | list(string) | yes | "Max Planck; Marie Curie" | names of the author(s) of this dataset |
| license | string | yes | "CC BY 4.0" | license for the distribution of the dataset. |
| doi | anyURI | yes | "10.5281/zenodo.596252" | A digital document identifier referencing the dataset already archived in a data repository |
license | License name | Reference | Comment |
|---|---|---|---|
"CC BY" | Creative commons | https://creativecommons.org | all flavours of the creative commons license scheme (e.g., CC BY-SA, CC BY-NC) |
"CC0" | Creative Commons Public Domain Dedication | https://creativecommons.org | you give up the copyright on your data and make it public domain. Please note this cannot be revoked! |
"Copyright" | Copyright protected | - | Data are copyright protected and cannot be used or distributed without the creator’s agreement |
3.5.2 Example
{...}
<xlum xmlns:xlum="xlum.org" lang="en"
formatVersion="1.0" flavour="generic" author="Marie Curie; Max Planck"
license="CC BY 4.0" doi="NA">
<sample ...>
<sequence ...>
<record ...>
<curve ...>
{...}
</curve>
{...}
</record>
</sequence>
{...}
</sample>
{...}
</xlum>
{...}3.6 General parameters (all nodes)
A few attributes are available on all node levels except the root node <xlum>. These attributes are mainly of technical nature to ease the sequential data storage during the measurement process (such as parentID).
| Identifier | Data type | Allows NA? | Example | Information |
|---|---|---|---|---|
| state | string | yes | "recording" | defines the state of the node; only used by equipment manufactures |
| parentID | token | yes | "201007145551910" | uniquely identifying the parent node, the root node has the ID 0, this is usually not needed but may help to store data |
| comment | String | yes | "what a wonderful sample" | comment field |
4 Special cases
This section provides suggestions for attributes for particular types of measurements. These attributes are not compulsory, but if needed should be used for the sake of consistency.
Please note: these metadata are not required to store and analyse the data!
4.1 Pulsing data (optional)
In measurements with pulsed light stimulation, data processing usually requires summations of photon counts on the hardware level using so-called field-programmable gate arrays (FPGA). It is neither meaningful nor feasible to store single counts in separate curves for photon arrival times of \(10^{-6} \leq \tau \leq 10^{-3}\) s. Hence, multiple detectors <curve/> data may be stored per <record/>, one for each (x) pulse(-s). In those cases, the <record/> node contains additional metadata that may help understand the data and pre-processing by the hardware level better. All attributes are optional.
| Identifier | Data type | Example | Information |
|---|---|---|---|
| onTime | numeric | "1E-01" | on time of stimulation per pulse, [s] |
| offTime | numeric | "1E-01" | on time of stimulation per pulse, [s] |
| nPulses | numeric | "10" | number of stimulation pulses total |
| summations | numeric | "2" | how many consecutive pulse records are summed (e.g. 10 pulses, 2 summations -> 5 records) |
| channelsPerPulse | numeric | "1000" | number of channels that are recorded in one curve |
| countsNormalised | numeric | "1" | if not 0, counts per channel are normalized to counts per second (helpful with uneven distributed channel times) |
Also the curves contain additional metadata
| Identifier | Data type | Example | Information |
|---|---|---|---|
| pulseID | numeric | "1" | current pulse number |
5 Practical examples
This section contains detailed examples for particular curve types in order to provide usable examples for the data format.
<?xml version="1.0" encoding="utf-8"?>
<xlum lang="en" version="1.0" flavour="xlum" doiRef="">
<sample ...>
<sequence ...>
<record ...>
<curve ...>
12 23 23 23 13 23
</curve>
</record>
</sequence>
</sample>
</xlum>6 Appendix
6.1 Risø BIN/BINX to XLUM metadata argument matching
Risø BIN/BINX files consist of a metadata header block and a data block. Some of the metadata serves the sole purpose of describing the structure of the data block, others return the specific equipment status but are non-relevant for the data analysis.
Most BIN/BINX metadata is covered by the XLUM format by design and hence finds their expression implicitly. The following argument list contains all possible arguments in BIN/BINX files, however, not all BIN/BINX file versions support all arguments. Covered here are BIN/BINX files between versions 03 to 08.
| # | BIN/BINX argument | Expression in XLUM | Remarks |
|---|---|---|---|
| 01 | VERSION | NONE | number of BIN/BINX file version |
| 02 | PREVIOUS | implicit | covered through XLUM structure |
| 03 | NPOINTS | implicit | can be calculated from data |
| 04 | RECTYPE | NONE | file format internal metadata |
| 05 | RUN | implicit | structure through order in <sequence/> |
| 06 | SET | implicit | structure through order in <sequence/> |
| 07 | POSITION | position | in <sequence/> node |
| 08 | GRAIN | implicit | a grain is considered a sample |
| 09 | GRAINNUMBER | implicit | a grain is considered a sample |
| 10 | CURVENO | implicit | through structure |
| 11 | XCOORD | NONE | considered non-relevant |
| 12 | YCOORD | NONE | considered non-relevant |
| 13 | SAMPLE | name | in <sample/> node |
| 14 | COMMENT | comment | allowed argument in every node |
| 15 | SYSTEMID | readerSN | in <sequence/> node |
| 16 | FNAME | fileName | in <sequence/> node |
| 17 | USER | author | in <xlum/> node |
| 18 | TIME | startDate | in <curve/> node (ISO 8601-1:2019) |
| 19 | DATE | startDate | in <curve/> node (ISO 8601-1:2019) |
| 20 | DTYPE | sampleCondition | in <curve/> node |
| 21 | BL_TIME | implicit | can be expressed as <curve/> |
| 22 | BL_UNIT | implicit | expressed in <curve/> metadata |
| 23 | NORM1 | implicit | should be part of <curve/> representation |
| 24 | NORM2 | implicit | should be part of <curve/> representation |
| 25 | NORM3 | implicit | should be part of <curve/> representation |
| 26 | BG | implicit | can be expressed as <curve/> |
| 27 | SHIFT | implicit | part of curve information |
| 28 | TAG | NONE | non-relevant internal metadata |
| 29 | LTYPE | recordType | in <record/> node |
| 30 | LIGHTSOURCE | implicit | can be added in comment fields |
| 31 | LPOWER | implicit | can be expressed as <curve/> |
| 32 | LIGHTPOWER | implicit | can be expressed as <curve/> |
| 33 | LOW | implicit | not needed, part of <curve/> |
| 34 | HIGH | implicit | not needed, part of <curve/> |
| 35 | RATE | implicit | not needed, part of <curve/> |
| 36 | TEMPERATURE | implicit | expressed as <curve/> |
| 37 | MEASTEMP | implicit | expressed as <curve/> |
| 38 | AN_TEMP | implicit | expressed as <curve/> |
| 39 | AN_TIME | implicit | expressed as <curve/> |
| 40 | TOLDELAY | implicit | expressed as <curve/> |
| 41 | TOLON | implicit | expressed as <curve/> |
| 42 | TOLOFF | implicit | expressed as <curve/> |
| 43 | IRR_TIME | implicit | expressed as <curve/> |
| 44 | IRR_TYPE | NONE | can be added as comment |
| 45 | IRR_UNIT | tUnit | in <curve/> node |
| 46 | IRR_DOSERATE | NONE | either as curve or comment |
| 47 | IRR_DOSERATEERR | NONE | only as comment possible |
| 48 | TIMESINCEIRR | implicit | calculated from time stamps |
| 49 | TIMETICK | implicit | derived from onTIME and offTIME |
| 50 | ONTIME | onTIME | in <record/> node |
| 51 | OFFTIME | offTIME | in <record/> node |
| 52 | STIMEPERIOD | implicit | through nPulses in <record/> node |
| 53 | GATE_ENABLED | NONE | considered non-relevant |
| 54 | ENABLE_FLAGS | NONE | considered non-relevant |
| 55 | GATE_START | NONE | considered non-relevant |
| 56 | GATE_STOP | NONE | considered non-relevant |
| 57 | PTENABLED | NONE | considered non-relevant |
| 58 | DTENABLED | NONE | considered non-relevant |
| 59 | DEADTIME | NONE | considered non-relevant |
| 60 | MAXLPOWER | implicit | represented as <curve/> |
| 61 | XRF_ACQTIME | implicit | represented as <curve/> |
| 62 | XRF_HV | implicit | represented as <curve/> |
| 63 | XRF_CURR | implicit | represented as <curve/> |
| 64 | XRF_DEADTIMEEF | NONE | possible as comment |
| 65 | DETECTOR_ID | component | in <curve/> node |
| 66 | LOWERFILTER_ID | filter | in <curve/> node |
| 67 | LOWERFILTER_ID | filter | in <curve/> node |
| 68 | ENOISEFACTOR | NONE | usage unknown |
| 69 | MARKPOS_X1 | implicit | can be expressed as <curve/> or comment |
| 70 | MARKPOS_Y1 | implicit | can be expressed as <curve/> or comment |
| 71 | MARKPOS_X2 | implicit | can be expressed as <curve/> or comment |
| 72 | MARKPOS_Y2 | implicit | can be expressed as <curve/> or comment |
| 73 | MARKPOS_X3 | implicit | can be expressed as <curve/> or comment |
| 74 | MARKPOS_Y3 | implicit | can be expressed as <curve/> or comment |
| 75 | EXTR_START | NONE | usage unknown |
| 76 | EXTR_END | NONE | usage unknown |
| 77 | SEQUENCE | name | in <sequence/> node |
Note: metadata tagged with “considered non-relevant” or “usage unknown” indicate that they are not considered (yet) as relevant enough for long-term data preservation and are hence not to be included as a compulsory argument in the XLUM format definition. However, they can still be stored in the XLUM format regardless.
6.2 Freiberg Instruments XSYG argument matching
The XYSG file format by Freiberg Instruments is already an XML-based file format . It can be somewhat considered the predecessor of the XLUM format. The structure was developed in February 2013 but never formally published, and attributes are not necessarily followed. The main differences are:
- The XSYG file format consists of only four nodes instead of five. Parent node
xlumexists only for the XLUM format - Nodes start with a capital letter (e.g.,
<Sample>, instead of<sample>in XLUM) - The individual curves values are stored as a pair-list with values separated by a semicolon
;, in the<Curve>node, i.e.,x0 , y0 ; x1 , y1 ; x2 , y2 ; x3 , y3. XLUM uses attributes instead, and values are stored separately through white space - For spectrometer data, the XSYG format defines an alternative storage format of values in the
<Curve>node of form1.0 ,[554|555|559|553|556|550| {...}. Contrary, XLUM maintains a consistent storage format. - The chosen date format was not ISO conform yyyyMMddhhmmss instead of ISO 8601-1:2019: YYYY-MM-DDThh:mm:ss[.mmm]Z recalculated to Zulu time.
In the following, the XYSG to XLUM argument matching is listed node-wise.
6.2.1 <Curve/> vs <curve/>
| # | XSYG attribute | Expression in XLUM | Remarks |
|---|---|---|---|
| 01 | startDate | startDate | note the format difference |
| 02 | duration | duration | |
| 03 | curveType | duration | |
| 04 | stimulator | component | |
| 05 | detector | component | |
| 06 | curveDscripter | xLabel, yLabel, tLabel, vLabel, xUnit, yUnit, tUnit, vUnit | needs to be split and matched separately |
| 07 | offset | implicit | can be encode through the tValues |
| 08 | detectionWindow | detectionWindow | |
| 09 | filterIDs | NONE | purely manufacture related ID |
| 10 | filterNames | filter | filter names separated by ; |
6.2.2 <Record/> vs <record/>
| # | XSYG attribute | Expression in XLUM | Remarks |
|---|---|---|---|
| 01 | startDate | startDate | note the format difference |
| 02 | endDate | implicit | can be calculated from other attributes |
| 03 | recordType | recordType | |
| 04 | name | NONE | encoded through recordType + comment |
| 05 | sequenceStepNumber | sequenceStepNumber | |
| 06 | metaIrrType | implicit | encode through component in <curve/> |
| 07 | metaIrrDuration | implicit | through <curve/> node |
| 08 | sampleCondition | sampleCondition |
6.2.3 <Sequence/> vs <sequence/>
| # | XSYG attribute | Expression in XLUM | Remarks |
|---|---|---|---|
| 01 | position | position | |
| 02 | creationDate | implicit | through startDate of <record/> and <curve/> |
| 03 | name | name | |
| 04 | protocol | implicit | in name |
| 05 | mineral | NONE | value to be entered in <sample/> node |
6.2.4 <Sample/> vs <sample/>
| # | XSYG attribute | Expression in XLUM | Remarks |
|---|---|---|---|
| 01 | name | name | |
| 02 | user | NONE | Author can be entered in <xlum/> node |
| 03 | startDate | implicit | through values in other <curve/> and <record/> nodes |
| 04 | sampleCarrier | NONE | entry in general attribute comment possible |
| 05 | lexsygID | implicit | entry in attributes in <sequence/> node attributes |
| 06 | lexStudioVersion | implicit | entry in attributes in <sequence/> node attributes |
| 07 | firmwareVersion | implicit | entry in attributes in <sequence/> node attributes |
| 08 | os | NONE | however, entry in comment attribute possible |
6.2.5 General attributes
| # | XSYG attribute | Expression in XLUM | Remarks |
|---|---|---|---|
| 01 | state | state | |
| 02 | partentID | partentID | |
| 03 | comment | comment |
6.3 SUERC PSL argument matching
PSL files are produced by portable luminescence readers produced by the Scottish Universities Environmental Research Centre (SUERC). PSL files are loosely structured ASCII files without format descriptions.
Measured values are stored in essentially five columns (three columns, but total count and count values per cycle include uncertainties), example:
Time (s) Total Count Counts per Cycle
-------- --------------------- ---------------------
1 0 +/- 4 0 +/- 4
2 -2 +/- 6 -2 +/- 4
3 12 +/- 8 14 +/- 5These values can be represented in the XLUM format using <curve/> nodes,
one <curve/> for the counts per cycle and another for the uncertainties.
This solution is safer than adding uncertainties as attributes and more consistent
because the uncertainties must have been estimated using a component.
Columns under “Total Count” represent cumulative data that can be recalculated from the values under “Counts per Cycle” should not be stored explicitly in XLUM.
Additionally, PSL provides arguments that can be extracted from those files. The matching listed below is non-exhaustive and represents the current state-of-knowledge about the PSL format.
| # | PSL metadata | Expression in XLUM | Remarks |
|---|---|---|---|
| 01 | Run Name | implicit | through comment |
| 02 | Sample no | name | through name in <sample/> node |
| 03 | Sequence name | name | through name in <sequence/> node |
| 04 | Filename | fileName | in <sequence/> node |
| 05 | Dark Count | implicit | as <curve/> node |
| 06 | Light Count | implicit | as <curve/> node |
| 07 | Dark Count Correction | NONE | this is a logical attribute, which can part of comment if needed in the <curve/> node |
| 08 | Offset Subract | NONE | this can be part of comment in <curve/> node |
| 09 | Datafile Path | NONE | not considered relevant |
| 10 | Summary Path | NONE | not considered relevant |
| 11 | Run Sequence | implicit | through name in <sequence/> node |
| 12 | Timestamp | startDate | in <curve/> node, please note the different format |
| 13 | Measurement | recordType | in <record/> node |
| 14 | Stim | recordType | in <record/> node |
| 15 | On/Off(us) | onTime, offTime | in <curve/> node |
| 16 | Cycle(ms), No | nPulses | in <curve/> node |
Please note: as for the data formats above, in principle, all arguments can be carried over directly. However, for the sake of consistency, the matching should be applied.