This function performs a luminescence spectra deconvolution on
RLum.Data.Spectrum and matrix objects on an energy scale.
The function is optimised for emission spectra typically obtained in the
context of TL, OSL and RF measurements detected between 200 and 1000 nm.
The function is not designed to deconvolve TL curves (counts against
temperature; no wavelength scale). If you are interested in such analysis,
please check, e.g., package 'tgcd'.
Usage
fit_EmissionSpectra(
object,
frame = NULL,
n_components = NULL,
start_parameters = NULL,
sub_negative = 0,
input_scale = NULL,
method_control = list(),
verbose = TRUE,
plot = TRUE,
...
)Arguments
- object
RLum.Data.Spectrum, matrix (required): input object. Please note that an energy spectrum is expected
- frame
integer (optional): number of the frame to be analysed. If
NULL, all available frames are analysed.- n_components
integer (optional): maximum number of components desired: the number of component actually fitted may be smaller than this. Can be combined with other parameters.
- start_parameters
numeric (optional): allows to provide own start parameters for a semi-automated procedure. Parameters need to be provided in eV. Every value provided replaces a value from the automated peak finding algorithm (in ascending order).
- sub_negative
numeric (with default): substitute negative values in the input object by the number provided here (default:
0). Can be set toNULL, i.e. negative values are kept.- input_scale
character (optional): defines whether your x-values are expressed as wavelength or energy values. Allowed values are
"wavelength","energy"orNULL, in which case the function tries to guess the input automatically.- method_control
list (optional): options to control the fit method and the output produced, see details.
- verbose
logical (with default): enable/disable output to the terminal.
- plot
logical (with default): enable/disable the plot output.
- ...
further arguments to be passed to control the plot output (supported:
main,xlab,ylab,xlim,ylim,log,mtext,legend(TRUEorFALSE),legend.text,legend.pos)
Value
———————————–[ NUMERICAL OUTPUT ]
———————————–
RLum.Results-object
slot: @data
| Element | Type | Description |
$data | matrix | the final fit matrix |
$fit | nls | the fit object returned by minpack.lm::nls.lm |
$fit_info | list | a few additional parameters that can be used to assess the quality of the fit |
$df_plot | list | values of all curves in the plot for each
frame analysed (only if method_control$export.plot.data = TRUE) |
slot: @info
The original function call
———————————[ TERMINAL OUTPUT ]
———————————
The terminal output provides brief information on the
deconvolution process and the obtained results.
Terminal output is only shown when verbose = TRUE.
—————————[ PLOT OUTPUT ]
—————————
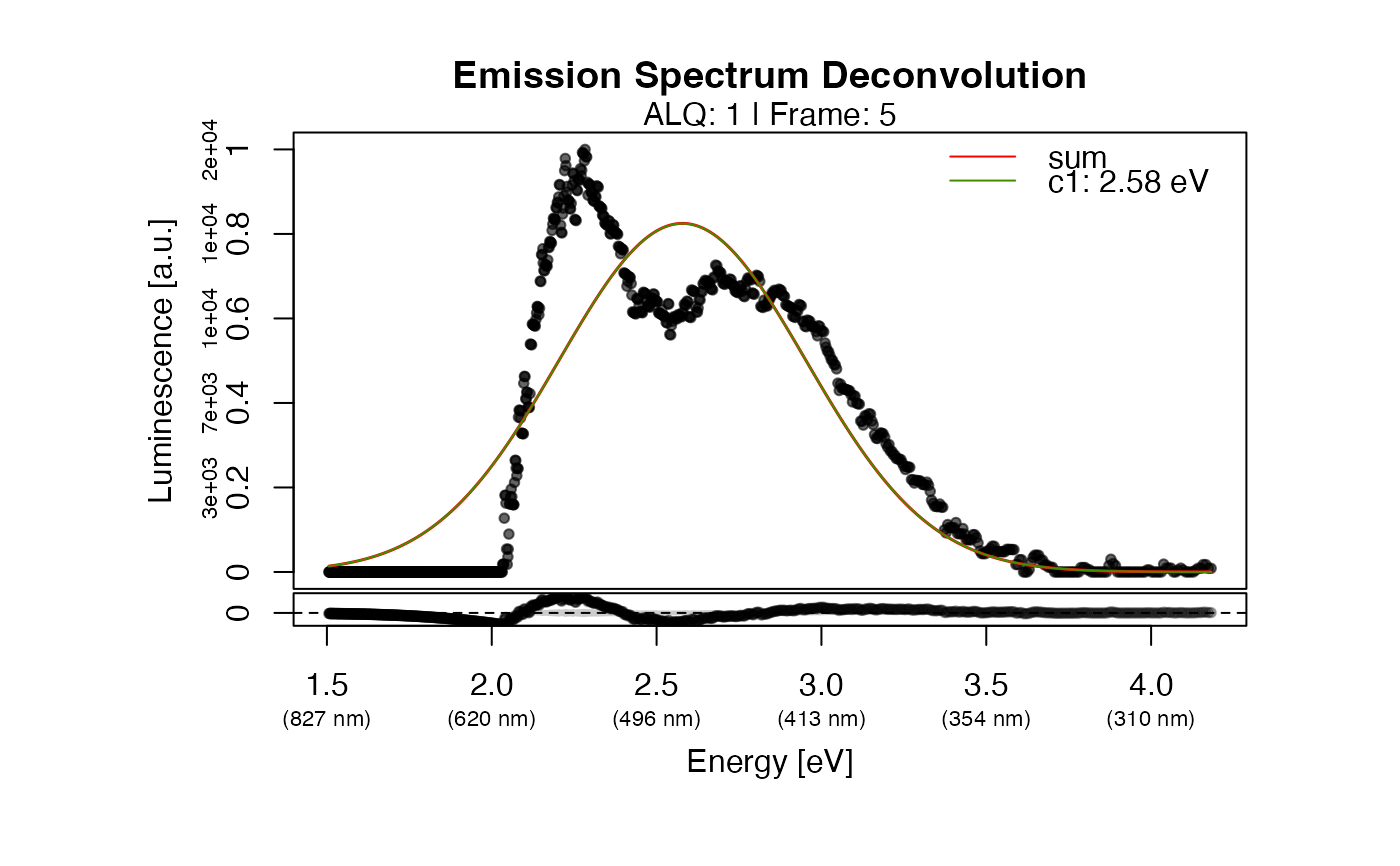
The function returns a plot showing the raw signal with the detected components. If the fitting failed, a basic plot is returned showing the raw data and indicating the peaks detected for the start parameter estimation. The grey band in the residual plot indicates the 10% deviation from 0 (means no residual).
Details
Used equation
The emission spectra (on an energy scale) can be best described as the sum of multiple Gaussian components:
'$$ y = \Sigma Ci * 1/(\sigma_{i} * \sqrt(2 * \pi)) * exp(-1/2 * ((x - \mu_{i})/\sigma_{i}))^2) $$
with the parameters \(\sigma\) (peak width) and \(\mu\) (peak centre) and \(C\) (scaling factor).
Start parameter estimation and fitting algorithm
The spectrum deconvolution consists of the following steps:
Peak finding
Start parameter estimation
Fitting via minpack.lm::nls.lm
The peak finding is realised by an approach (re-)suggested by Petr Pikal via the R-help
mailing list (https://stat.ethz.ch/pipermail/r-help/2005-November/thread.html) in November 2005.
This goes back to even earlier discussion in 2001 based on Prof Brian Ripley's idea.
It smartly uses the functions stats::embed and max.col to identify peaks positions.
For the use in this context, the algorithm has been further modified to scale on the
input data resolution (cf. source code).
The start parameter estimation uses random sampling from a range of meaningful parameters
and repeats the fitting until 1000 successful fits have been produced or the set max.runs value
is exceeded.
Currently the best fit is the one with the lowest number for squared residuals, but
other parameters are returned as well. If a series of curves needs to be analysed,
it is recommended to make few trial runs, then fix the number of components and
run at least 10,000 iterations (parameter method_control = list(max.runs = 10000)).
Supported method_control settings
| Parameter | Type | Default | Description |
max.runs | integer | 10000 | maximum allowed search iterations, if exceed the searching stops |
graining | numeric | 15 | control over how coarse or fine the spectrum is split into search intervals for the peak finding algorithm |
norm | logical | TRUE | normalise data to the highest count value before fitting |
export.plot.data | logical | FALSE | enable/disable export of the values of all curves in the plot for each frame analysed |
trace | logical | FALSE | enable/disable the tracing of the minimisation routine |
Author
Sebastian Kreutzer, Institute of Geography, Heidelberg University (Germany)
Marco Colombo, Institute of Geography, Heidelberg University (Germany)
, RLum Developer Team
How to cite
Kreutzer, S., Colombo, M., 2025. fit_EmissionSpectra(): Luminescence Emission Spectra Deconvolution. Function version 0.1.3. In: Kreutzer, S., Burow, C., Dietze, M., Fuchs, M.C., Schmidt, C., Fischer, M., Friedrich, J., Mercier, N., Philippe, A., Riedesel, S., Autzen, M., Mittelstrass, D., Gray, H.J., Galharret, J., Colombo, M., Steinbuch, L., Boer, A.d., 2025. Luminescence: Comprehensive Luminescence Dating Data Analysis. R package version 1.1.2. https://r-lum.github.io/Luminescence/
Examples
##load example data
data(ExampleData.XSYG, envir = environment())
##subtract background
TL.Spectrum@data <- TL.Spectrum@data[] - TL.Spectrum@data[,15]
results <- fit_EmissionSpectra(
object = TL.Spectrum,
frame = 5,
method_control = list(max.runs = 10)
)
#>
#> [fit_EmissionSpectra()]
#>
#> >> Treating dataset >> 5 <<
#> >> Wavelength scale detected ...
#> >> Wavelength to energy scale conversion ... [OK]
#>
>> Searching components ... [-]
>> Searching components ... [\]
>> Searching components ... [-]
>> Searching components ... [-]
>> Searching components ... [/]
>> Searching components ... [-]
>> Searching components ... [/]
>> Searching components ... [-]
>> Searching components ... [\]
>> Searching components ... [-]
>> Searching components ... [OK]
#>
#> >> Fitting results (1 component model):
#> -------------------------------------------------------------------------
#> mu SE(mu) sigma SE(sigma) C SE(C)
#> [1,] 2.578403 0.006164002 0.3748351 0.005834795 0.7750779 0.0108509
#> -------------------------------------------------------------------------
#> SE: standard error | SSR: 2.164e+01| R^2: 0.807 | R^2_adj: 0.1938
#> (use the output in $fit for a more detailed analysis)
#>
 ##deconvolution of a TL spectrum
if (FALSE) { # \dontrun{
##load example data
##replace 0 values
results <- fit_EmissionSpectra(
object = TL.Spectrum,
frame = 5, main = "TL spectrum"
)
} # }
##deconvolution of a TL spectrum
if (FALSE) { # \dontrun{
##load example data
##replace 0 values
results <- fit_EmissionSpectra(
object = TL.Spectrum,
frame = 5, main = "TL spectrum"
)
} # }