The function computes De value and De value error using the global standardised growth curve (gSGC) assumption proposed by Li et al., 2015 for OSL dating of sedimentary quartz.
Usage
calc_gSGC(
data,
gSGC.type = "0-250",
gSGC.parameters = NULL,
n.MC = 100,
verbose = TRUE,
plot = TRUE,
...
)Arguments
- data
data.frame (required): input data the following columns five columns in the given order:
LnTn,LnTn.error,Lr1Tr1,Lr1Tr1.error,Dr1. Column names are not required.- gSGC.type
character (with default): function parameters to use for the iteration procedure, either
"0-450"or"0-250", as presented in Li et al., 2015 (Table 2). This is ignored ifgSGC.parametersis set.- gSGC.parameters
list (optional): option to provide own function parameters used for fitting as named list. Nomenclature follows Li et al., 2015, i.e.
list(A, A.error, D0, D0.error, c, c.error, Y0, Y0.error, range), whererangeis defines the interval where the function is considered as valid, e.g.range = c(0,250).
If set, optiongSGC.typewill be ignored.- n.MC
integer (with default): number of Monte Carlo simulation runs for error estimation, see details.
- verbose
logical (with default): enable/disable output to the terminal.
- plot
logical (with default): enable/disable the plot output.
- ...
parameters will be passed to the plot output
Value
Returns an S4 object of type RLum.Results.
@data$ De.value (data.frame) .. $ De .. $ De.error .. $ Eta $ De.MC (list) contains the matrices from the error estimation.$ uniroot (list) contains the uniroot outputs of the De estimations
@info
`$ call“ (call) the original function call
Details
The error of the De value is determined using a Monte Carlo simulation approach.
Solving of the equation is realised using uniroot.
Large values for n.MC will significantly increase the computation time.
How to cite
Kreutzer, S., 2025. calc_gSGC(): Calculate De value based on the gSGC by Li et al., 2015. Function version 0.1.3. In: Kreutzer, S., Burow, C., Dietze, M., Fuchs, M.C., Schmidt, C., Fischer, M., Friedrich, J., Mercier, N., Philippe, A., Riedesel, S., Autzen, M., Mittelstrass, D., Gray, H.J., Galharret, J., Colombo, M., Steinbuch, L., Boer, A.d., 2025. Luminescence: Comprehensive Luminescence Dating Data Analysis. R package version 1.1.2. https://r-lum.github.io/Luminescence/
References
Li, B., Roberts, R.G., Jacobs, Z., Li, S.-H., 2015. Potential of establishing a 'global standardised growth curve' (gSGC) for optical dating of quartz from sediments. Quaternary Geochronology 27, 94-104. doi:10.1016/j.quageo.2015.02.011
Author
Sebastian Kreutzer, Institute of Geography, Heidelberg University (Germany) , RLum Developer Team
Examples
results <- calc_gSGC(data = data.frame(
LnTn = 2.361, LnTn.error = 0.087,
Lr1Tr1 = 2.744, Lr1Tr1.error = 0.091,
Dr1 = 34.4))
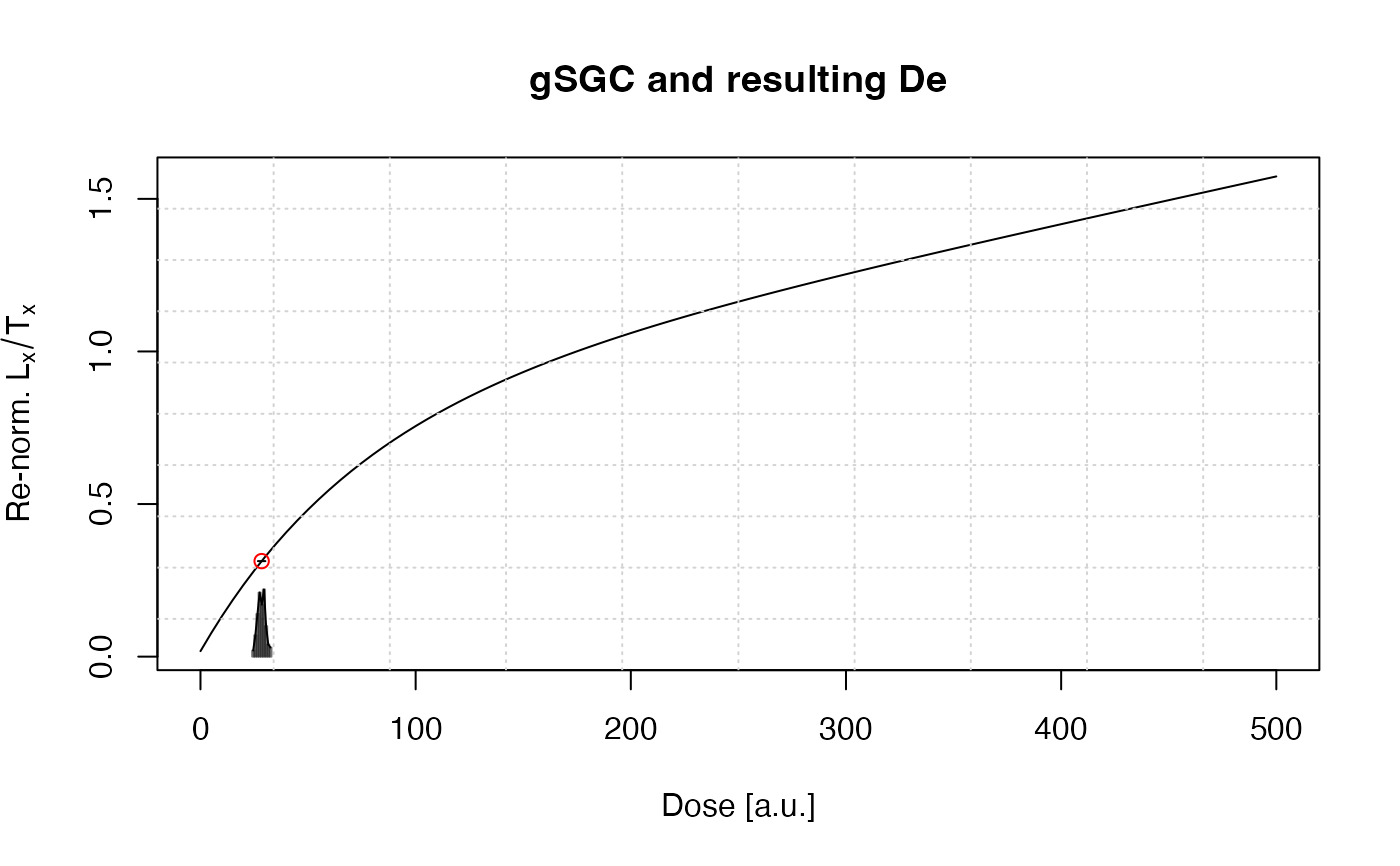 #>
#> [calc_gSGC()]
#> Corresponding De based on the gSGC
#>
#> Ln/Tn: 2.361 ± 0.087
#> Lr1/Tr1: 2.744 ± 0.091
#> Dr1: 34.4
#> f(D): 0.787 * (1 - exp(-D /73.9)) + c * D + 0.01791
#> n.MC: 100
#> ------------------------------
#> De: 28.43 ± 1.5
#> ------------------------------
get_RLum(results, data.object = "De")
#> DE DE.ERROR ETA
#> 1 28.42881 1.495121 0.1325632
#>
#> [calc_gSGC()]
#> Corresponding De based on the gSGC
#>
#> Ln/Tn: 2.361 ± 0.087
#> Lr1/Tr1: 2.744 ± 0.091
#> Dr1: 34.4
#> f(D): 0.787 * (1 - exp(-D /73.9)) + c * D + 0.01791
#> n.MC: 100
#> ------------------------------
#> De: 28.43 ± 1.5
#> ------------------------------
get_RLum(results, data.object = "De")
#> DE DE.ERROR ETA
#> 1 28.42881 1.495121 0.1325632