Run Monte-Carlo Simulation for CW-OSL (delocalized transitions)
Source:R/run_MC_CW_OSL_DELOC.R
run_MC_CW_OSL_DELOC.RdRuns a Monte-Carlo (MC) simulation of continuous wave optically stimulated luminescence (CW-OSL) using the one trap one recombination centre (OTOR) model. The term delocalized here refers to the involvement of the conduction band.
Usage
run_MC_CW_OSL_DELOC(
A,
times,
clusters = 10,
N_e = 200,
n_filled = N_e,
R,
method = "par",
output = "signal",
...
)Arguments
- A
numeric (required): The optical excitation rate from trap to conduction band (s^-1)
- times
numeric (required): The sequence of temperature steps within the simulation (s)
- clusters
numeric (with default): The number of created clusters for the MC runs. The input can be the output of create_ClusterSystem. In that case
n_filledindicate absolute numbers of a system.- N_e
integer (with default): The total number of electron traps available (dimensionless). Can be a vector of
length(clusters), shorter values are recycled.- n_filled
integer (with default): The number of filled electron traps at the beginning of the simulation (dimensionless). Can be a vector of
length(clusters), shorter values are recycled.- R
numeric (required): The retrapping ratio for delocalized transitions (dimensionless)
- method
character (with default): Sequential
'seq'or parallel'par'processing. In the parallel mode the function tries to run the simulation on multiple CPU cores (if available) with a positive effect on the computation time.- output
character (with default): Output is either the
'signal'(the default) or'remaining_e'(the remaining charges, electrons, in the trap)- ...
further arguments, such as
coresto control the number of used CPU cores orverboseto silence the terminal
Value
This function returns an object of class RLumCarlo_Model_Output which
is a list consisting of an array with dimension length(times) x clusters
and a numeric time vector.
Details
The model
$$ I_{DELOC}(t) = -dn/dt = A * (n^2 / (N*R + n(1-R))) $$
Where in the function:
t := time (s)
A := the optical excitation rate from trap to conduction band (1/s)
n := n_filled, the instantaneous number of electrons
N := N_e the available number of electron traps available
R := retrapping ratio for delocalized transitions
How to cite
Kreutzer, S., 2025. run_MC_CW_OSL_DELOC(): Run Monte-Carlo Simulation for CW-OSL (delocalized transitions). Function version 0.1.0. In: Friedrich, J., Kreutzer, S., Pagonis, V., Schmidt, C., 2025. RLumCarlo: Monte-Carlo Methods for Simulating Luminescence Phenomena. R package version 0.1.10. https://r-lum.github.io/RLumCarlo/
References
Pagonis, V., Friedrich, J., Discher, M., Müller-Kirschbaum, A., Schlosser, V., Kreutzer, S., Chen, R. and Schmidt, C., 2019. Excited state luminescence signals from a random distribution of defects: A new Monte Carlo simulation approach for feldspar. Journal of Luminescence 207, 266–272. doi:10.1016/j.jlumin.2018.11.024
Further reading
Chen, R., McKeever, S.W.S., 1997. Theory of Thermoluminescence and Related Phenomena. WORLD SCIENTIFIC. doi:10.1142/2781
Examples
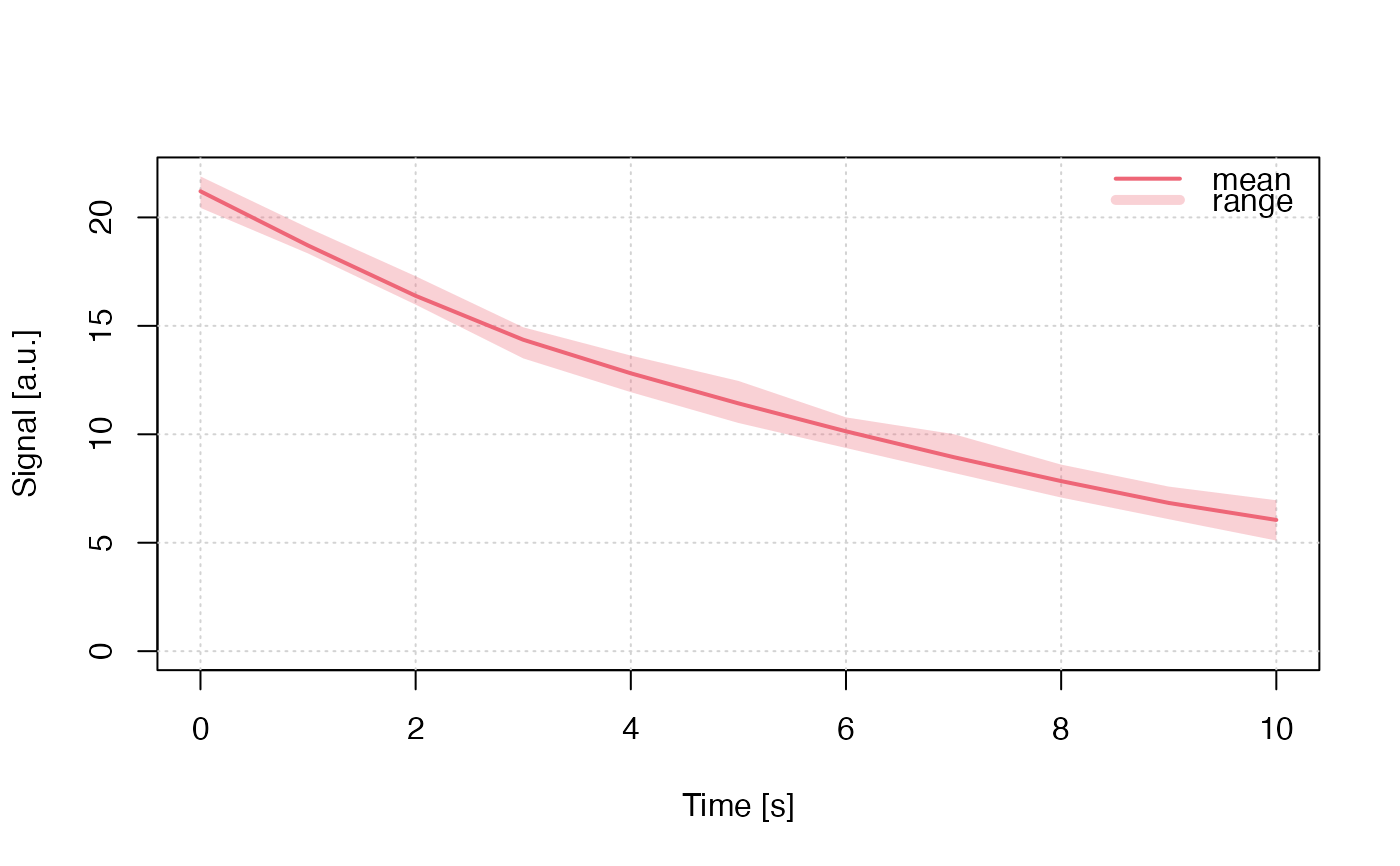
## brief example
run_MC_CW_OSL_DELOC(
A = 0.12,
R = 0.1,
times = 0:10,
clusters = 10,
method = "seq") %>%
plot_RLumCarlo(legend = TRUE)
 ## A long example
if (FALSE) { # \dontrun{
A <- c(0.1,0.3,0.5,1)
times <- seq(0, 60, 1)
s <- 1e12
E <- 1
R <- c(1e-7, 1e-6, 0.01, 0.1) # sequence of different R values
clusters <- 1000 # number of Monte Carlo simulations
N_e <- c(200, 500, 700, 400) # number of free electrons
n_filled <- c(200, 500, 100, 70) # number of filled traps
method <-"par"
output <- "signal"
col <- c(1,2,3,4) # ifferent colours for the individual curves
plot_uncertainty <- c(TRUE,FALSE,TRUE,FALSE) # do you want to see the uncertainty?
add_TF <- c(FALSE,rep(TRUE, (length(R)-1)))
## loop to plot different curves into one plot
for (u in 1:length(R)){
results <- run_MC_CW_OSL_DELOC(
A = A[u],
times,
clusters = clusters,
N_e = N_e[u],
n_filled = n_filled[u],
R = R[u],
method = method,
output = output)
plot_RLumCarlo(
results,
add = add_TF[u],
legend = FALSE,
col = col[u],
main = "Delocalised Transition")
}
# add your legend with your parameters
legend("topright",
ncol = 4,
cex = 0.55,
title = "parameters",
legend=c(
paste0("A = ", A),
paste0("n_filled = ", n_filled),
paste0("N_e = ", N_e),
paste0("R = ", R)),
bty = "n",
text.col = col)
} # }
## A long example
if (FALSE) { # \dontrun{
A <- c(0.1,0.3,0.5,1)
times <- seq(0, 60, 1)
s <- 1e12
E <- 1
R <- c(1e-7, 1e-6, 0.01, 0.1) # sequence of different R values
clusters <- 1000 # number of Monte Carlo simulations
N_e <- c(200, 500, 700, 400) # number of free electrons
n_filled <- c(200, 500, 100, 70) # number of filled traps
method <-"par"
output <- "signal"
col <- c(1,2,3,4) # ifferent colours for the individual curves
plot_uncertainty <- c(TRUE,FALSE,TRUE,FALSE) # do you want to see the uncertainty?
add_TF <- c(FALSE,rep(TRUE, (length(R)-1)))
## loop to plot different curves into one plot
for (u in 1:length(R)){
results <- run_MC_CW_OSL_DELOC(
A = A[u],
times,
clusters = clusters,
N_e = N_e[u],
n_filled = n_filled[u],
R = R[u],
method = method,
output = output)
plot_RLumCarlo(
results,
add = add_TF[u],
legend = FALSE,
col = col[u],
main = "Delocalised Transition")
}
# add your legend with your parameters
legend("topright",
ncol = 4,
cex = 0.55,
title = "parameters",
legend=c(
paste0("A = ", A),
paste0("n_filled = ", n_filled),
paste0("N_e = ", N_e),
paste0("R = ", R)),
bty = "n",
text.col = col)
} # }