The function performs a SAR TL analysis on a RLum.Analysis object including growth curve fitting.
Arguments
- object
RLum.Analysis or a list of such objects (required) : input object containing data for analysis
- object.background
currently not used
- signal.integral.min
integer (required): requires the channel number for the lower signal integral bound (e.g.
signal.integral.min = 100)- signal.integral.max
integer (required): requires the channel number for the upper signal integral bound (e.g.
signal.integral.max = 200)- integral_input
character (with default): defines the input for the arguments
signal.integral.minandsignal.integral.max. These limits can be either provided'channel'number (the default) or'temperature'. If'temperature'is chosen, the best matching channel is selected.- sequence.structure
vector character (with default): specifies the general sequence structure. Three steps are allowed (
"PREHEAT","SIGNAL","BACKGROUND"), in addition a parameter"EXCLUDE". This allows excluding TL curves which are not relevant for the protocol analysis. (Note: No TL are removed by default)- rejection.criteria
list (with default): list containing rejection criteria in percentage for the calculation.
- dose.points
numeric (optional): option set dose points manually
- log
character (with default): a character string which contains
"x"if the x-axis is to be logarithmic,"y"if the y axis is to be logarithmic and"xy"or"yx"if both axes are to be logarithmic. See plot.default).- ...
further arguments that will be passed to the function fit_DoseResponseCurve
Value
A plot (optional) and an RLum.Results object is returned containing the following elements:
- De.values
data.frame containing De-values and further parameters
- LnLxTnTx.values
data.frame of all calculated
Lx/Txvalues including signal, background counts and the dose points.- rejection.criteria
data.frame with values that might by used as rejection criteria. NA is produced if no R0 dose point exists.
The output should be accessed using the function get_RLum.
Details
This function performs a SAR TL analysis on a set of curves. The SAR
procedure in general is given by Murray and Wintle (2000). For the
calculation of the Lx/Tx value the function calc_TLLxTxRatio is
used.
Provided rejection criteria
[recycling.ratio]: calculated for every repeated regeneration dose point.
[recuperation.rate]: recuperation rate calculated by
comparing the Lx/Tx values of the zero regeneration point with the Ln/Tn
value (the Lx/Tx ratio of the natural signal). For methodological
background see Aitken and Smith (1988)
Note
THIS IS A BETA VERSION
No TL curves will be removed from the input object without further warning.
How to cite
Kreutzer, S., 2025. analyse_SAR.TL(): Analyse SAR TL measurements. Function version 0.3.1. In: Kreutzer, S., Burow, C., Dietze, M., Fuchs, M.C., Schmidt, C., Fischer, M., Friedrich, J., Mercier, N., Philippe, A., Riedesel, S., Autzen, M., Mittelstrass, D., Gray, H.J., Galharret, J., Colombo, M., Steinbuch, L., Boer, A.d., 2025. Luminescence: Comprehensive Luminescence Dating Data Analysis. R package version 1.1.2. https://r-lum.github.io/Luminescence/
References
Aitken, M.J. and Smith, B.W., 1988. Optical dating: recuperation after bleaching. Quaternary Science Reviews 7, 387-393.
Murray, A.S. and Wintle, A.G., 2000. Luminescence dating of quartz using an improved single-aliquot regenerative-dose protocol. Radiation Measurements 32, 57-73.
Author
Sebastian Kreutzer, Institute of Geography, Heidelberg University (Germany) , RLum Developer Team
Examples
##load data
data(ExampleData.BINfileData, envir = environment())
##transform the values from the first position in a RLum.Analysis object
object <- Risoe.BINfileData2RLum.Analysis(TL.SAR.Data, pos=3)
##perform analysis
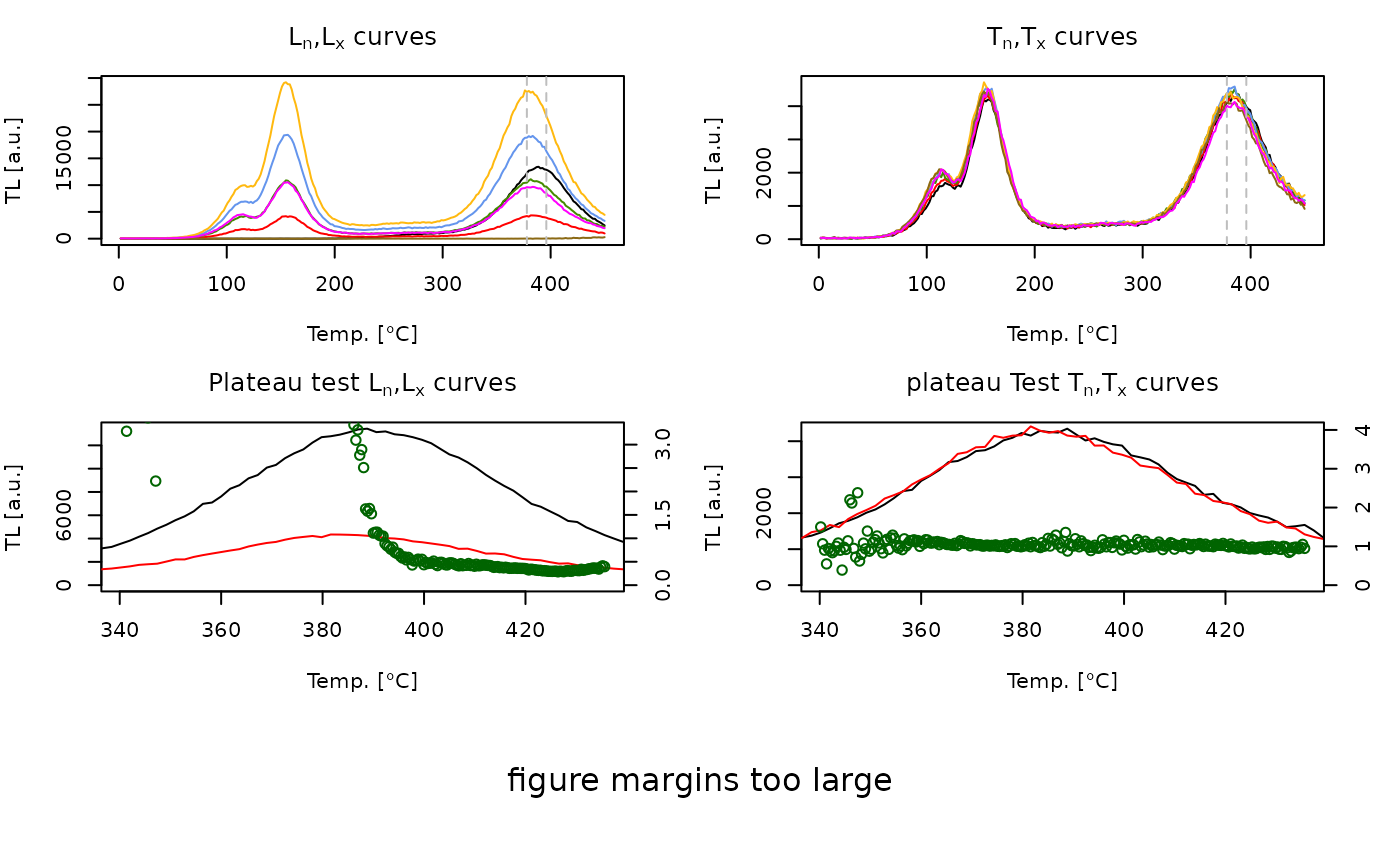
analyse_SAR.TL(
object = object,
signal.integral.min = 210,
signal.integral.max = 220,
fit.method = "EXP OR LIN",
sequence.structure = c("SIGNAL", "BACKGROUND"))
 #> [fit_DoseResponseCurve()] Fit: EXP OR LIN (interpolation) | De = 415.66 | D01 = 3685556.6
#>
#> [RLum.Results-class]
#> originator: analyse_SAR.TL()
#> data: 3
#> .. $data : data.frame
#> .. $LnLxTnTx.table : data.frame
#> .. $rejection.criteria : data.frame
#> additional info elements: 1
#> [fit_DoseResponseCurve()] Fit: EXP OR LIN (interpolation) | De = 415.66 | D01 = 3685556.6
#>
#> [RLum.Results-class]
#> originator: analyse_SAR.TL()
#> data: 3
#> .. $data : data.frame
#> .. $LnLxTnTx.table : data.frame
#> .. $rejection.criteria : data.frame
#> additional info elements: 1