Apply the central age model (CAM) after Galbraith et al. (1999) to a given De distribution
Source:R/calc_CentralDose.R
calc_CentralDose.RdThis function calculates the central dose and dispersion of the De
distribution, their standard errors and the profile log likelihood function
for sigma.
This function uses the equations of Galbraith & Roberts (2012). The
parameters delta and sigma are estimated by numerically solving
eq. 15 and 16. Their standard errors are approximated using eq. 17.
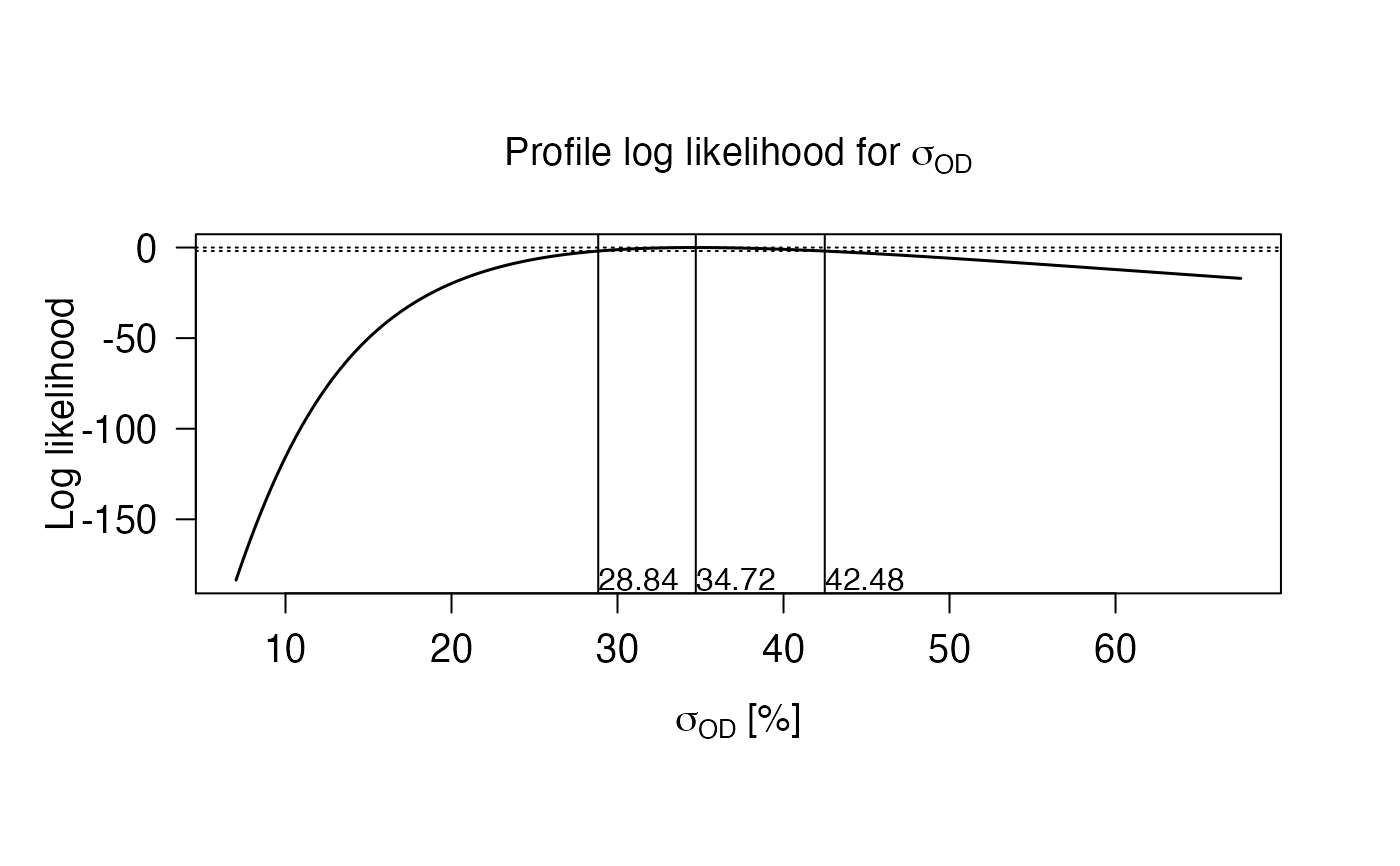
In addition, the profile log-likelihood function for sigma is
calculated using eq. 18 and presented as a plot. Numerical values of the
maximum likelihood approach are only presented in the plot and not
in the console. A detailed explanation on maximum likelihood estimation can
be found in the appendix of Galbraith & Laslett (1993, 468-470) and
Galbraith & Roberts (2012, 15)
Arguments
- data
RLum.Results or data.frame (required): for data.frame: two columns with De
(data[,1])and De error(data[,2]). Records containing missing values will be removed.- sigmab
numeric (with default): additional spread in De values, representing the expected overdispersion in the data should the sample be well-bleached (Cunningham & Wallinga 2012, p. 100). NOTE: For the logged model (
log = TRUE) this value must be a fraction, e.g. 0.2 (= 20 %). If the un-logged model is used (log = FALSE), sigmab must be provided in the same absolute units of the De values (seconds or Gray).- log
logical (with default): fit the (un-)logged central age model to De data. Log transformation is allowed only if the De values are positive.
- plot
logical (with default): enable/disable the plot output.
- ...
further arguments (
trace,verbose).
Value
Returns a plot (optional) and terminal output. In addition an RLum.Results object is returned containing the following elements:
- .$summary
data.frame summary of all relevant model results.
- .$data
data.frame original input data
- .$args
list used arguments
- .$call
call the function call
- .$profile
data.frame the log likelihood profile for sigma
The output should be accessed using the function get_RLum.
How to cite
Burow, C., 2025. calc_CentralDose(): Apply the central age model (CAM) after Galbraith et al. (1999) to a given De distribution. Function version 1.5. In: Kreutzer, S., Burow, C., Dietze, M., Fuchs, M.C., Schmidt, C., Fischer, M., Friedrich, J., Mercier, N., Philippe, A., Riedesel, S., Autzen, M., Mittelstrass, D., Gray, H.J., Galharret, J., Colombo, M., Steinbuch, L., Boer, A.d., 2025. Luminescence: Comprehensive Luminescence Dating Data Analysis. R package version 1.1.2. https://r-lum.github.io/Luminescence/
References
Galbraith, R.F. & Laslett, G.M., 1993. Statistical models for mixed fission track ages. Nuclear Tracks Radiation Measurements 4, 459-470.
Galbraith, R.F., Roberts, R.G., Laslett, G.M., Yoshida, H. & Olley, J.M., 1999. Optical dating of single grains of quartz from Jinmium rock shelter, northern Australia. Part I: experimental design and statistical models. Archaeometry 41, 339-364.
Galbraith, R.F. & Roberts, R.G., 2012. Statistical aspects of equivalent dose and error calculation and display in OSL dating: An overview and some recommendations. Quaternary Geochronology 11, 1-27.
Further reading
Arnold, L.J. & Roberts, R.G., 2009. Stochastic modelling of multi-grain equivalent dose (De) distributions: Implications for OSL dating of sediment mixtures. Quaternary Geochronology 4, 204-230.
Bailey, R.M. & Arnold, L.J., 2006. Statistical modelling of single grain quartz De distributions and an assessment of procedures for estimating burial dose. Quaternary Science Reviews 25, 2475-2502.
Cunningham, A.C. & Wallinga, J., 2012. Realizing the potential of fluvial archives using robust OSL chronologies. Quaternary Geochronology 12, 98-106.
Rodnight, H., Duller, G.A.T., Wintle, A.G. & Tooth, S., 2006. Assessing the reproducibility and accuracy of optical dating of fluvial deposits. Quaternary Geochronology, 1 109-120.
Rodnight, H., 2008. How many equivalent dose values are needed to obtain a reproducible distribution?. Ancient TL 26, 3-10.
Author
Christoph Burow, University of Cologne (Germany)
Based on a rewritten S script of Rex Galbraith, 2010
, RLum Developer Team
Examples
##load example data
data(ExampleData.DeValues, envir = environment())
##apply the central dose model
calc_CentralDose(ExampleData.DeValues$CA1)
#>
#> [calc_CentralDose]
#>
#> ----------- meta data ----------------
#> n: 62
#> log: TRUE
#> ----------- dose estimate ------------
#> abs. central dose: 65.71
#> abs. SE: 3.05
#> rel. SE [%]: 4.65
#> ----------- overdispersion -----------
#> abs. OD: 22.79
#> abs. SE: 2.27
#> OD [%]: 34.69
#> SE [%]: 3.46
#> -------------------------------------
#>