(Un-)logged minimum age model (MAM) after Galbraith et al. (1999)
Source:R/calc_MinDose.R
calc_MinDose.RdFunction to fit the (un-)logged three or four parameter minimum dose model (MAM-3/4) to De data.
Parameters
This model has four parameters:
gamma: | minimum dose on the log scale |
mu: | mean of the non-truncated normal distribution |
sigma: | spread in ages above the minimum |
p0: | proportion of grains at gamma |
If par=3 (default) the 3-parameter minimum age model is applied,
where gamma=mu. For par=4 the 4-parameter model is applied instead.
(Un-)logged model
In the original version of the minimum dose model, the basic data are the natural
logarithms of the De estimates and relative standard errors of the De
estimates. The value for sigmab must be provided as a ratio
(e.g, 0.2 for 20 %). This model will be applied if log = TRUE.
If log=FALSE, the modified un-logged model will be applied instead. This
has essentially the same form as the original version. gamma and
sigma are in Gy and gamma becomes the minimum true dose in the
population.
Note that the un-logged model requires sigmab to be in the same
absolute unit as the provided De values (seconds or Gray).
While the original (logged) version of the minimum dose model may be appropriate for most samples (i.e. De distributions), the modified (un-logged) version is specially designed for modern-age and young samples containing negative, zero or near-zero De estimates (Arnold et al. 2009, p. 323).
Initial values & boundaries
The log-likelihood calculations use the nlminb function for box-constrained
optimisation using PORT routines. Accordingly, initial values for the four
parameters can be specified via init.values. If no values are
provided for init.values, reasonable starting values are estimated
from the input data. If the final estimates of gamma, mu,
sigma and p0 are totally off target, consider providing custom
starting values via init.values.
The boundaries for the individual model parameters are not required to be
explicitly specified. If you want to override the default boundary values,
use arguments gamma.lower, gamma.upper, sigma.lower, sigma.upper,
p0.lower, p0.upper, mu.lower and mu.upper.
Bootstrap
When bootstrap=TRUE the function applies the bootstrapping method as
described in Cunningham & Wallinga (2012). By default, the minimum age model
produces 1000 first level and 3000 second level bootstrap replicates. The
uncertainty on sigmab is 0.04 by default. These values can be changed by
using the arguments bs.M (first level replicates), bs.N
(second level replicates) and sigmab.sd (error on sigmab). The bandwidth
of the kernel density estimate can be specified with bs.h. By default,
this is calculated as:
$$h = (2*\sigma_{DE})/\sqrt{n}$$
Multicore support
Parallel computations can be activated by setting multicore=TRUE.
By default, the number of available logical CPU cores is determined
automatically, but can be changed with cores. The multicore support
is only available when bootstrap=TRUE and spawns n R instances
for each core to get MAM estimates for each of the N and M bootstrap
replicates. Note that this option is highly experimental and may or may not
work for your machine. The performance gain increases for larger number
of bootstrap replicates. However, note that with each additional core (and
hence R instance) the memory usage can significantly increase, depending
on the number of bootstrap replicates. When insufficient memory is available,
there will be a massive performance hit.
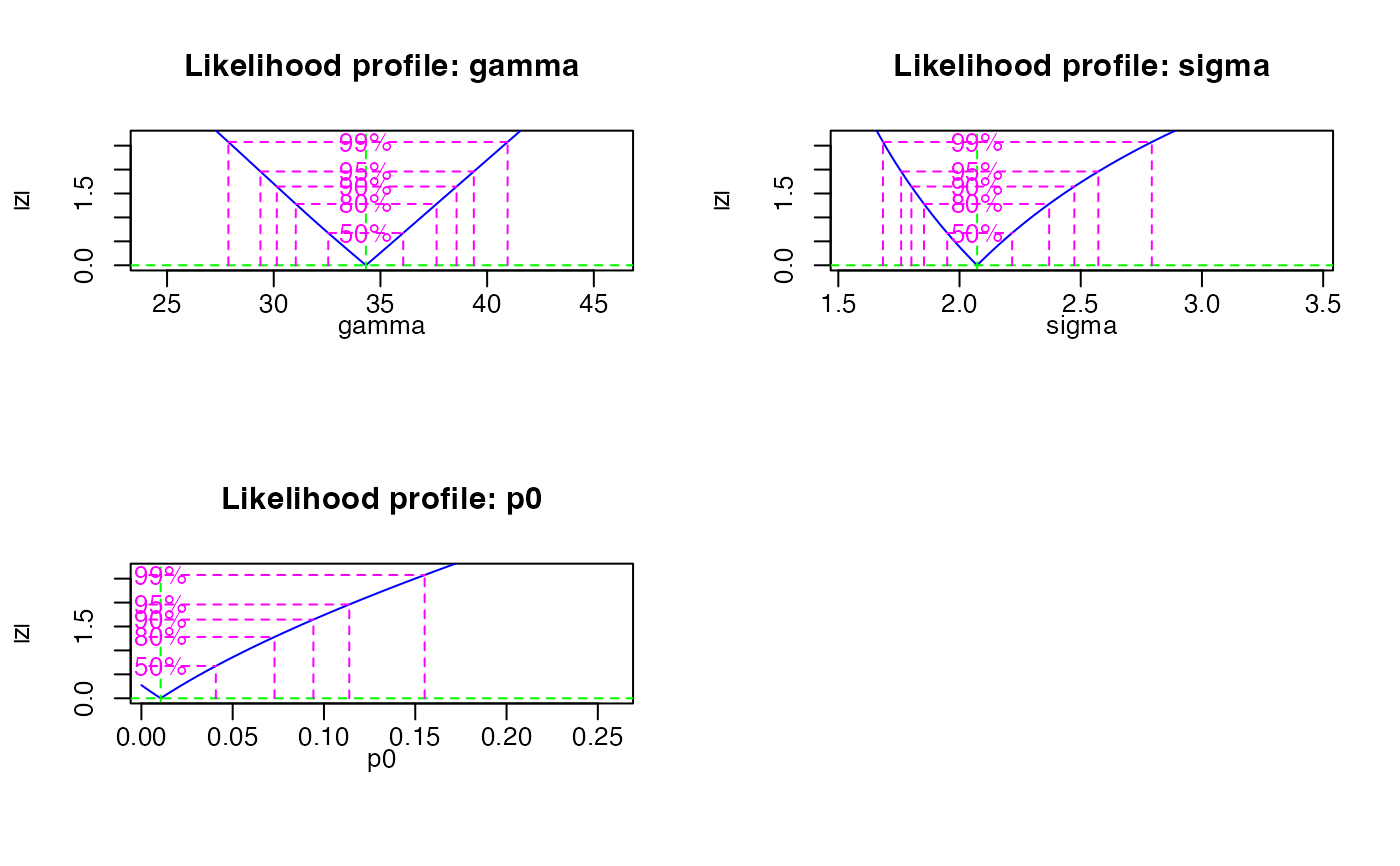
Likelihood profiles
The likelihood profiles are generated and plotted by the bbmle package.
The profile likelihood plots look different to ordinary profile likelihood as
"[...] the plot method for likelihood profiles displays the square root of
the deviance difference (twice the difference in negative log-likelihood from
the best fit), so it will be V-shaped for cases where the quadratic approximation
works well [...]." (Bolker 2016).
For more details on the profile likelihood
calculations and plots please see the vignettes of the bbmle package
(also available here: https://CRAN.R-project.org/package=bbmle.
Usage
calc_MinDose(
data,
sigmab,
log = TRUE,
par = 3,
bootstrap = FALSE,
init.values = NULL,
level = 0.95,
log.output = FALSE,
plot = TRUE,
multicore = FALSE,
...
)Arguments
- data
RLum.Results or data.frame (required): for data.frame: two columns for De and De error.
- sigmab
numeric (required): additional spread in De values, representing the expected overdispersion in the data should the sample be well-bleached (Cunningham & Wallinga 2012, p. 100). NOTE: For the logged model (
log = TRUE) this value must be a fraction, e.g. 0.2 (= 20 %). If the un-logged model is used (log = FALSE),sigmabmust be provided in the same absolute units of the De values (seconds or Gray). See details.- log
logical (with default): whether the logged minimum dose model should be fit to De data.
- par
numeric (with default): number of parameters in the minimum age model, either 3 (default) or 4.
- bootstrap
logical (with default): apply the recycled bootstrap approach of Cunningham & Wallinga (2012).
- init.values
numeric (optional): a named list with starting values for
gamma,sigma,p0andmu(e.g.list(gamma=100, sigma=1.5, p0=0.1, mu=100)). If no values are provided, reasonable values will be estimated from the data. NOTE: the initial values must always be given in the absolute units. If a logged model is applied (log = TRUE), the providedinit.valuesare automatically log-transformed.- level
logical (with default): the confidence level required (defaults to 0.95).
- log.output
logical (with default): if
TRUEandlog = TRUE, the console output will also show the logged values of the final parameter estimates and confidence intervals.- plot
logical (with default): enable/disable the plot output.
- multicore
logical (with default): parallelize the computation of the bootstrap by creating a multicore cluster (only considered if
bootstrap = TRUE). By default, it uses all available logical CPU cores, but this can be changed with optioncores. Note that this option is highly experimental and may not work on all machines.- ...
(optional) further arguments for bootstrapping (
bs.M,bs.N,bs.h,sigmab.sd). See details for their usage. Further arguments areverbose: enable/disable output to the terminaldebug: enable/disable extended console outputcores: number of cores to be used whenmulticore=TRUE
Value
Returns a plot (optional) and terminal output. In addition an RLum.Results object is returned containing the following elements:
- $summary
data.frame summary of all relevant model results.
- $data
data.frame original input data
- $args
list used arguments
- $call
call the function call
- $mle
bbmle::mle2 object containing the maximum log likelihood functions for all parameters
- $BIC
numeric BIC score
- $confint
data.frame confidence intervals for all parameters
- $profile
stats::profile the log likelihood profiles
- $bootstrap
list bootstrap results
The output should be accessed using the function get_RLum.
Note
The default starting values for gamma, mu, sigma
and p0 may only be appropriate for some De data sets and may need to
be changed for other data. This is especially true when the un-logged
version is applied.
Also note that all R warning messages are suppressed
when running this function. If the results seem odd consider re-running the
model with debug=TRUE which provides extended console output and
forwards all internal warning messages.
How to cite
Burow, C., 2025. calc_MinDose(): (Un-)logged minimum age model (MAM) after Galbraith et al. (1999). Function version 0.4.6. In: Kreutzer, S., Burow, C., Dietze, M., Fuchs, M.C., Schmidt, C., Fischer, M., Friedrich, J., Mercier, N., Philippe, A., Riedesel, S., Autzen, M., Mittelstrass, D., Gray, H.J., Galharret, J., Colombo, M., Steinbuch, L., Boer, A.d., 2025. Luminescence: Comprehensive Luminescence Dating Data Analysis. R package version 1.1.2. https://r-lum.github.io/Luminescence/
References
Arnold, L.J., Roberts, R.G., Galbraith, R.F. & DeLong, S.B., 2009. A revised burial dose estimation procedure for optical dating of young and modern-age sediments. Quaternary Geochronology 4, 306-325.
Galbraith, R.F. & Laslett, G.M., 1993. Statistical models for mixed fission track ages. Nuclear Tracks Radiation Measurements 4, 459-470.
Galbraith, R.F., Roberts, R.G., Laslett, G.M., Yoshida, H. & Olley, J.M., 1999. Optical dating of single grains of quartz from Jinmium rock shelter, northern Australia. Part I: experimental design and statistical models. Archaeometry 41, 339-364.
Galbraith, R.F., 2005. Statistics for Fission Track Analysis, Chapman & Hall/CRC, Boca Raton.
Galbraith, R.F. & Roberts, R.G., 2012. Statistical aspects of equivalent dose and error calculation and display in OSL dating: An overview and some recommendations. Quaternary Geochronology 11, 1-27.
Olley, J.M., Roberts, R.G., Yoshida, H., Bowler, J.M., 2006. Single-grain optical dating of grave-infill associated with human burials at Lake Mungo, Australia. Quaternary Science Reviews 25, 2469-2474.
Further reading
Arnold, L.J. & Roberts, R.G., 2009. Stochastic modelling of multi-grain equivalent dose (De) distributions: Implications for OSL dating of sediment mixtures. Quaternary Geochronology 4, 204-230.
Bolker, B., 2016. Maximum likelihood estimation analysis with the bbmle package. In: Bolker, B., R Development Core Team, 2016. bbmle: Tools for General Maximum Likelihood Estimation. R package version 1.0.18. https://CRAN.R-project.org/package=bbmle
Bailey, R.M. & Arnold, L.J., 2006. Statistical modelling of single grain quartz De distributions and an assessment of procedures for estimating burial dose. Quaternary Science Reviews 25, 2475-2502.
Cunningham, A.C. & Wallinga, J., 2012. Realizing the potential of fluvial archives using robust OSL chronologies. Quaternary Geochronology 12, 98-106.
Rodnight, H., Duller, G.A.T., Wintle, A.G. & Tooth, S., 2006. Assessing the reproducibility and accuracy of optical dating of fluvial deposits. Quaternary Geochronology 1, 109-120.
Rodnight, H., 2008. How many equivalent dose values are needed to obtain a reproducible distribution?. Ancient TL 26, 3-10.
Author
Christoph Burow, University of Cologne (Germany)
Based on a rewritten S script of Rex Galbraith, 2010
The bootstrap approach is based on a rewritten MATLAB script of Alastair Cunningham.
Alastair Cunningham is thanked for his help in implementing and cross-checking the code.
, RLum Developer Team
Examples
## Load example data
data(ExampleData.DeValues, envir = environment())
# (1) Apply the minimum age model with minimum required parameters.
# By default, this will apply the un-logged 3-parameter MAM.
calc_MinDose(data = ExampleData.DeValues$CA1, sigmab = 0.1)
#>
#> ----------- meta data -----------
#> n par sigmab logged Lmax BIC
#> 62 3 0.1 TRUE -43.57969 106.4405
#>
#> --- final parameter estimates ---
#> gamma sigma p0 mu
#> 34.32 2.07 0.01 0
#>
#> ------ confidence intervals -----
#> 2.5 % 97.5 %
#> gamma 29.38 39.38
#> sigma 1.76 2.57
#> p0 NA 0.11
#>
#> ------ De (asymmetric error) -----
#> De lower upper
#> 34.32 29.38 39.38
#>
#> ------ De (symmetric error) -----
#> De error
#> 34.32 2.55
 #>
#> [RLum.Results-class]
#> originator: calc_MinDose()
#> data: 9
#> .. $summary : data.frame
#> .. $data : data.frame
#> .. $args : list
#> .. $call : call
#> .. $mle : mle2
#> .. $BIC : numeric
#> .. $confint : data.frame
#> .. $profile : profile.mle2
#> .. $bootstrap : list
#> additional info elements: 0
if (FALSE) { # \dontrun{
# (2) Re-run the model, but save results to a variable and turn
# plotting of the log-likelihood profiles off.
mam <- calc_MinDose(
data = ExampleData.DeValues$CA1,
sigmab = 0.1,
plot = FALSE)
# Show structure of the RLum.Results object
mam
# Show summary table that contains the most relevant results
res <- get_RLum(mam, "summary")
res
# Plot the log likelihood profiles retroactively, because before
# we set plot = FALSE
plot_RLum(mam)
# Plot the dose distribution in an abanico plot and draw a line
# at the minimum dose estimate
plot_AbanicoPlot(data = ExampleData.DeValues$CA1,
main = "3-parameter Minimum Age Model",
line = mam,polygon.col = "none",
hist = TRUE,
rug = TRUE,
summary = c("n", "mean", "mean.weighted", "median", "in.ci"),
centrality = res$de,
line.col = "red",
grid.col = "none",
line.label = paste0(round(res$de, 1), "\U00B1",
round(res$de_err, 1), " Gy"),
bw = 0.1,
ylim = c(-25, 18),
summary.pos = "topleft",
mtext = bquote("Parameters: " ~
sigma[b] == .(get_RLum(mam, "args")$sigmab) ~ ", " ~
gamma == .(round(log(res$de), 1)) ~ ", " ~
sigma == .(round(res$sig, 1)) ~ ", " ~
rho == .(round(res$p0, 2))))
# (3) Run the minimum age model with bootstrap
# NOTE: Bootstrapping is computationally intensive
# (3.1) run the minimum age model with default values for bootstrapping
calc_MinDose(data = ExampleData.DeValues$CA1,
sigmab = 0.15,
bootstrap = TRUE)
# (3.2) Bootstrap control parameters
mam <- calc_MinDose(data = ExampleData.DeValues$CA1,
sigmab = 0.15,
bootstrap = TRUE,
bs.M = 300,
bs.N = 500,
bs.h = 4,
sigmab.sd = 0.06,
plot = FALSE)
# Plot the results
plot_RLum(mam)
# save bootstrap results in a separate variable
bs <- get_RLum(mam, "bootstrap")
# show structure of the bootstrap results
str(bs, max.level = 2, give.attr = FALSE)
# print summary of minimum dose and likelihood pairs
summary(bs$pairs$gamma)
# Show polynomial fits of the bootstrap pairs
bs$poly.fits$poly.three
# Plot various statistics of the fit using the generic plot() function
par(mfcol=c(2,2))
plot(bs$poly.fits$poly.three, ask = FALSE)
# Show the fitted values of the polynomials
summary(bs$poly.fits$poly.three$fitted.values)
} # }
#>
#> [RLum.Results-class]
#> originator: calc_MinDose()
#> data: 9
#> .. $summary : data.frame
#> .. $data : data.frame
#> .. $args : list
#> .. $call : call
#> .. $mle : mle2
#> .. $BIC : numeric
#> .. $confint : data.frame
#> .. $profile : profile.mle2
#> .. $bootstrap : list
#> additional info elements: 0
if (FALSE) { # \dontrun{
# (2) Re-run the model, but save results to a variable and turn
# plotting of the log-likelihood profiles off.
mam <- calc_MinDose(
data = ExampleData.DeValues$CA1,
sigmab = 0.1,
plot = FALSE)
# Show structure of the RLum.Results object
mam
# Show summary table that contains the most relevant results
res <- get_RLum(mam, "summary")
res
# Plot the log likelihood profiles retroactively, because before
# we set plot = FALSE
plot_RLum(mam)
# Plot the dose distribution in an abanico plot and draw a line
# at the minimum dose estimate
plot_AbanicoPlot(data = ExampleData.DeValues$CA1,
main = "3-parameter Minimum Age Model",
line = mam,polygon.col = "none",
hist = TRUE,
rug = TRUE,
summary = c("n", "mean", "mean.weighted", "median", "in.ci"),
centrality = res$de,
line.col = "red",
grid.col = "none",
line.label = paste0(round(res$de, 1), "\U00B1",
round(res$de_err, 1), " Gy"),
bw = 0.1,
ylim = c(-25, 18),
summary.pos = "topleft",
mtext = bquote("Parameters: " ~
sigma[b] == .(get_RLum(mam, "args")$sigmab) ~ ", " ~
gamma == .(round(log(res$de), 1)) ~ ", " ~
sigma == .(round(res$sig, 1)) ~ ", " ~
rho == .(round(res$p0, 2))))
# (3) Run the minimum age model with bootstrap
# NOTE: Bootstrapping is computationally intensive
# (3.1) run the minimum age model with default values for bootstrapping
calc_MinDose(data = ExampleData.DeValues$CA1,
sigmab = 0.15,
bootstrap = TRUE)
# (3.2) Bootstrap control parameters
mam <- calc_MinDose(data = ExampleData.DeValues$CA1,
sigmab = 0.15,
bootstrap = TRUE,
bs.M = 300,
bs.N = 500,
bs.h = 4,
sigmab.sd = 0.06,
plot = FALSE)
# Plot the results
plot_RLum(mam)
# save bootstrap results in a separate variable
bs <- get_RLum(mam, "bootstrap")
# show structure of the bootstrap results
str(bs, max.level = 2, give.attr = FALSE)
# print summary of minimum dose and likelihood pairs
summary(bs$pairs$gamma)
# Show polynomial fits of the bootstrap pairs
bs$poly.fits$poly.three
# Plot various statistics of the fit using the generic plot() function
par(mfcol=c(2,2))
plot(bs$poly.fits$poly.three, ask = FALSE)
# Show the fitted values of the polynomials
summary(bs$poly.fits$poly.three$fitted.values)
} # }