The function performs a SAR CW-OSL analysis on an RLum.Analysis object, including growth curve fitting.
Usage
analyse_SAR.CWOSL(
object,
signal.integral.min = NA,
signal.integral.max = NA,
background.integral.min = NA,
background.integral.max = NA,
OSL.component = NULL,
rejection.criteria = list(),
dose.points = NULL,
dose.points.test = NULL,
trim_channels = FALSE,
mtext.outer = "",
plot = TRUE,
plot_onePage = FALSE,
plot_singlePanels = FALSE,
onlyLxTxTable = FALSE,
...
)Arguments
- object
RLum.Analysis (required): input object containing data for analysis, alternatively a list of RLum.Analysis objects can be provided. The object should only contain curves considered part of the SAR protocol (see Details).
- signal.integral.min
integer (required): lower bound of the signal integral. It can be a list of integers, if
objectis a list. If the input is a vector (e.g.,c(1,2)), the second value will be interpreted as the minimum signal integral for theTxcurve. It can be set toNA, in which case no integrals are taken into account.- signal.integral.max
integer (required): upper bound of the signal integral. It can be a list of integers if
objectis a list. If the input is a vector (e.g.,c(1,2)), the second value will be interpreted as the maximum signal integral for theTxcurve. It can be set toNA, in which case no integrals are taken into account.- background.integral.min
integer (required): lower bound of the background integral. It can be a list of integers if
objectis a list. If the input is a vector (e.g.,c(1,2)), the second value will be interpreted as the minimum background integral for theTxcurve. It can be set toNA, in which case no integrals are taken into account.- background.integral.max
integer (required): upper bound of the background integral. It can be a list of integers if
objectis a list. If the input is a vector (e.g.,c(1,2)), the second value will be interpreted as the maximum background integral for theTxcurve. It can be set toNA, in which case no integrals are taken into account.- OSL.component
character or integer (optional): single index or a character defining the signal component to be evaluated. It requires that the object was processed by
OSLdecomposition::RLum.OSL_decomposition. This argument can either be the name of the OSL component assigned byOSLdecomposition::RLum.OSL_global_fittingor the index in the descending order of decay rates. Then"1"selects the fastest decaying component,"2"the second fastest and so on. Can be a list of integers or strings (or mixed) If object is a list and this parameter is provided as list it alternates over the elements (aliquots) of the object list, e.g.,list(1,2)processes the first aliquot with component1and the second aliquot with component2.NULLdoes not process any component.- rejection.criteria
list (with default): provide a named list and set rejection criteria in percentage for further calculation. Can be a list in a list, if
objectis of type list. Note: If an unnamed list is provided the new settings are ignored!Allowed arguments are
recycling.ratio,recuperation.rate,palaeodose.error,testdose.error,exceed.max.regpoint = TRUE/FALSE,recuperation_reference("Natural" or any other dose point, e.g.,"R1"). Example:rejection.criteria = list(recycling.ratio = 10). By default, all numerical values are set to 10,exceed.max.regpoint = TRUE. Every criterion can be set toNA, in which case values are calculated, but they are not considered, i.e. their corresponding RC.Status is always'OK'.- dose.points
numeric (optional): a numeric vector containing the dose points values. Using this argument overwrites dose point values extracted from other data. Can be a list of numeric vectors, if
objectis of type list.- dose.points.test
numeric (optional): a numeric vector containing the test dose in the same units as
dose.points. If length = 1, the values will be recycled. It has only an effect forfit.method = 'OTORX'.- trim_channels
logical (with default): trim channels per record category to the lowest number of channels in the category by using trim_RLum.Data. Applies only to
OSLandIRSLcurves. For a more granular control use trim_RLum.Data before calling this function.- mtext.outer
character (optional): option to provide an outer margin
mtext. Can be a list of characters, ifobjectis of type list- plot
logical (with default): enable/disable the plot output.
- plot_onePage
logical (with default): enable/disable one page plot output.
- plot_singlePanels
logical (with default) or numeric (optional): single plot output (
TRUE/FALSE) to allow for plotting the results in single plot windows. If a numeric vector is provided the plots can be selected individually, i.e.plot_singlePanels = c(1,2,3,4)will plot the TL and Lx, Tx curves but not the legend (5) or the growth curve (6), (7) and (8) belong to rejection criteria plots. Requiresplot = TRUE.- onlyLxTxTable
logical (with default): If
TRUEthe dose response curve fitting and plotting is skipped. This allows to get hands on theLx/Txtable for large datasets without the need for a curve fitting.- ...
further arguments that will be passed to the functions fit_DoseResponseCurve, plot_DoseResponseCurve or calc_OSLLxTxRatio (supported:
background.count.distribution,sigmab,sig0). Please note that if you consider to use the early light subtraction method you should provide your ownsigmabvalue!
Value
A plot (optional) and an RLum.Results object is returned containing the following elements:
- data
data.frame containing De-values, De-error and further parameters
- LnLxTnTx.values
data.frame of all calculated Lx/Tx values including signal, background counts and the dose points
- rejection.criteria
data.frame with values that might by used as rejection criteria.
NAis produced if no R0 dose point exists.- Formula
formula formula that have been used for the growth curve fitting
The output should be accessed using the function get_RLum.
The function currently does support only 'OSL', 'IRSL' and 'POSL' data!
Details
The function performs an analysis for a standard SAR protocol measurements
introduced by Murray and Wintle (2000) with CW-OSL curves. For the
calculation of the Lx/Tx value the function calc_OSLLxTxRatio is
used. To change the way the Lx/Tx error is calculated use arguments
background.count.distribution and sigmab, which will be passed to
calc_OSLLxTxRatio.
What is part of a SAR sequence?
The function is rather picky when it comes down to accepted curve input (OSL, IRSL,...) and structure. A SAR sequence is basically a set of \(L_{x}/T_{x}\) curves. Hence, every second curve is considered a shine-down curve related to the test dose. It also means that the number of curves for \(L_{x}\) has to be equal to the number of \(T_{x}\) curves, and that hot-bleach curves do not belong into a SAR sequence; at least not for the analysis. Other curves allowed and processed are preheat curves, or preheat curves measured as TL, and irradiation curves. The later one indicates the duration of the irradiation, the dose and test dose points, e.g., as part of XSYG files.
Argument object is of type list
If the argument object is of type list containing only
RLum.Analysis objects, the function re-calls itself on each element
in the list. This is useful if to analyse an entire measurement without
writing separate for-loops. To gain in full control of the parameters (e.g., dose.points) for
every aliquot (corresponding to one RLum.Analysis object in the list), in
this case the arguments can be provided as list. This list should
be of similar length as the list provided with the argument object,
otherwise the function will create an own list of the requested length.
Function output will be just one single RLum.Results object.
Please be careful when using this option. While it may allow for a fast and efficient data analysis, the function may break with an unclear error message if the input data is misspecified.
Working with IRSL data
The function was originally designed to work just for 'OSL' curves, following the principles of the SAR protocol. An IRSL measurement protocol may follow this procedure, e.g., post-IR IRSL protocol (Thomsen et al., 2008). Therefore this functions has been enhanced to work with IRSL data, however, the function is only capable of analysing curves that follow the SAR protocol structure, i.e., to analyse a post-IR IRSL protocol, curve data have to be pre-selected by the user to fit the standards of the SAR protocol, i.e., Lx,Tx,Lx,Tx and so on.
Example: Imagine the measurement contains pIRIR50 and pIRIR225 IRSL
curves. Only one curve type can be analysed at the same time: either the
pIRIR50 curves or the pIRIR225 curves.
Supported rejection criteria
[recycling.ratio]: calculated for every repeated regeneration dose point.
[recuperation.rate]: recuperation rate calculated by comparing the
Lx/Tx values of the zero regeneration point with the Ln/Tn value (the
Lx/Tx ratio of the natural signal). For methodological background see
Aitken and Smith (1988). As a variant, recuperation_reference can be
specified to select another dose point as reference instead of Ln/Tn.
[testdose.error]: set the allowed error for the test dose, which by
default should not exceed 10%. The test dose error is calculated as
Tx_net.error/Tx_net. The calculation of the \(T_{n}\) error is detailed
in calc_OSLLxTxRatio.
[palaeodose.error]: set the allowed error for the De value, which per
default should not exceed 10%.
Irradiation times
The function makes two attempts to extra irradiation data (dose points)
automatically from the input object, if the argument dose.points is not
set (aka set to NULL).
It searches in every curve for an info object called
IRR_TIME. If this is found, any value set there is taken as dose point.If the object contains curves of type
irradiation, the function tries to use this information to assign these values to the curves. However, the function does not overwrite values preset inIRR_TIME.
How to cite
Kreutzer, S., 2025. analyse_SAR.CWOSL(): Analyse SAR CW-OSL Measurements. Function version 0.11.1. In: Kreutzer, S., Burow, C., Dietze, M., Fuchs, M.C., Schmidt, C., Fischer, M., Friedrich, J., Mercier, N., Philippe, A., Riedesel, S., Autzen, M., Mittelstrass, D., Gray, H.J., Galharret, J., Colombo, M., Steinbuch, L., Boer, A.d., 2025. Luminescence: Comprehensive Luminescence Dating Data Analysis. R package version 1.1.2. https://r-lum.github.io/Luminescence/
References
Aitken, M.J. and Smith, B.W., 1988. Optical dating: recuperation after bleaching. Quaternary Science Reviews 7, 387-393.
Duller, G., 2003. Distinguishing quartz and feldspar in single grain luminescence measurements. Radiation Measurements, 37 (2), 161-165.
Murray, A.S. and Wintle, A.G., 2000. Luminescence dating of quartz using an improved single-aliquot regenerative-dose protocol. Radiation Measurements 32, 57-73.
Thomsen, K.J., Murray, A.S., Jain, M., Boetter-Jensen, L., 2008. Laboratory fading rates of various luminescence signals from feldspar-rich sediment extracts. Radiation Measurements 43, 1474-1486. doi:10.1016/j.radmeas.2008.06.002
Author
Sebastian Kreutzer, Institute of Geography, Heidelberg University (Germany) , RLum Developer Team
Examples
##load data
##ExampleData.BINfileData contains two BINfileData objects
##CWOSL.SAR.Data and TL.SAR.Data
data(ExampleData.BINfileData, envir = environment())
##transform the values from the first position in a RLum.Analysis object
object <- Risoe.BINfileData2RLum.Analysis(CWOSL.SAR.Data, pos=1)
##perform SAR analysis and set rejection criteria
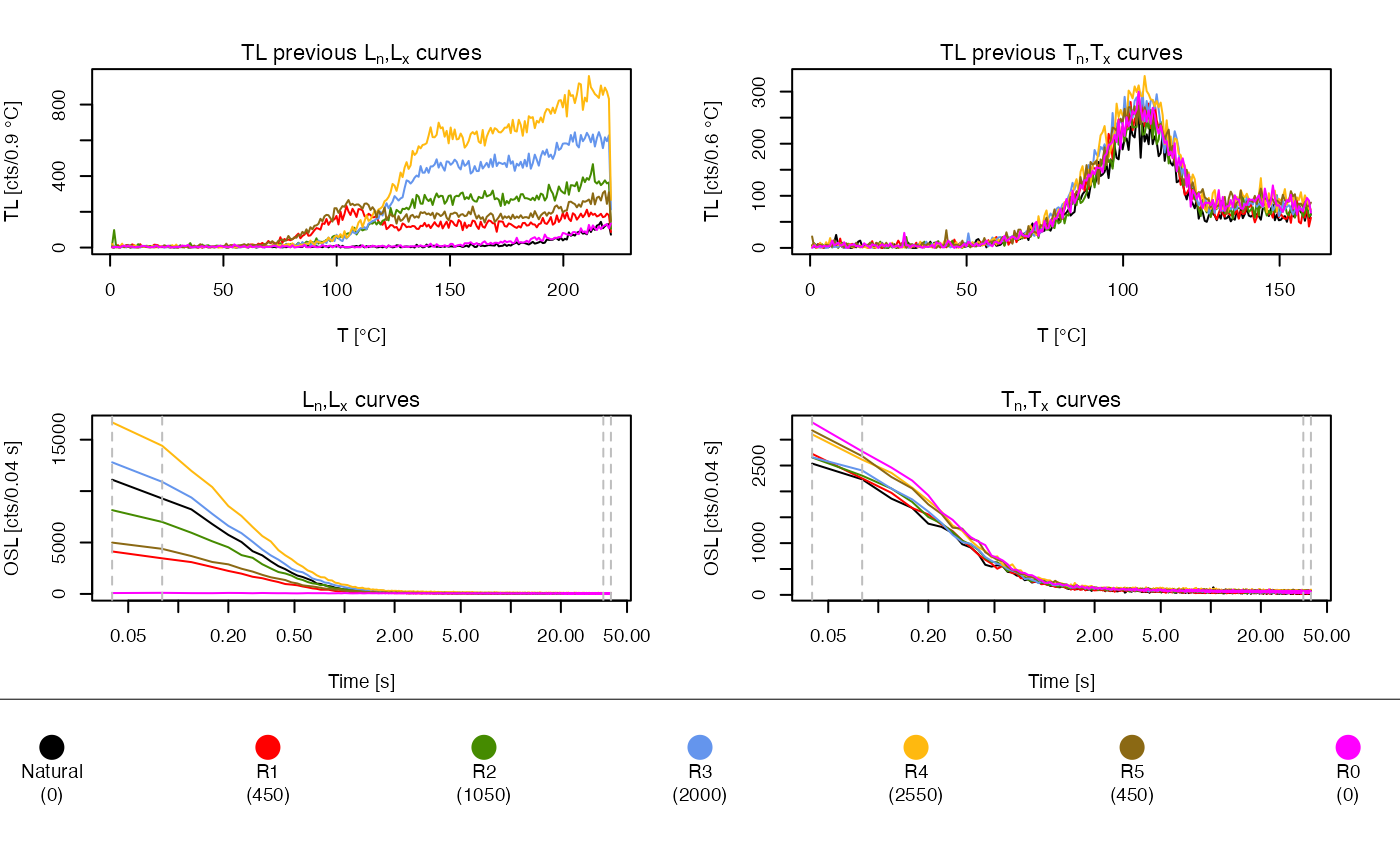
results <- analyse_SAR.CWOSL(
object = object,
signal.integral.min = 1,
signal.integral.max = 2,
background.integral.min = 900,
background.integral.max = 1000,
log = "x",
fit.method = "EXP",
plot_onePage = TRUE,
rejection.criteria = list(
recycling.ratio = 10,
recuperation.rate = 10,
testdose.error = 10,
palaeodose.error = 10,
recuperation_reference = "Natural",
exceed.max.regpoint = TRUE)
)
#> [fit_DoseResponseCurve()] Fit: EXP (interpolation) | De = 1668.25 | D01 = 1982.76
 ##show De results
get_RLum(results)
#> De De.Error D01 D01.ERROR D02 D02.ERROR Dc D63 n_N De.MC
#> 1 1668.251 46.78683 1982.757 107.7542 NA NA NA NA 0.4854696 1664.089
#> Fit Mode HPDI68_L HPDI68_U HPDI95_L HPDI95_U .De.plot .De.raw
#> 1 EXP interpolation 1611.046 1708.69 1570.575 1763.294 1668.251 1668.251
#> RC.Status signal.range background.range signal.range.Tx background.range.Tx
#> 1 OK 1:2 900:1000 NA:NA NA:NA
#> ALQ POS GRAIN UID
#> 1 1 1 0 5708d0a6ba7b4f22
##show LnTnLxTx table
get_RLum(results, data.object = "LnLxTnTx.table")
#> Name Repeated Dose LnLx LnLx.BG TnTx TnTx.BG Net_LnLx
#> 1 Natural FALSE 0 20391 60.15842 4771 73.50495 20330.84158
#> 2 R1 FALSE 450 7591 65.70297 4977 81.56436 7525.29703
#> 3 R2 FALSE 1050 15150 96.17822 4960 101.92079 15053.82178
#> 4 R3 FALSE 2000 23700 118.49505 5064 121.06931 23581.50495
#> 5 R4 FALSE 2550 31094 155.92079 5715 145.84158 30938.07921
#> 6 R5 TRUE 450 9376 122.37624 5860 127.60396 9253.62376
#> 7 R0 FALSE 0 192 94.39604 6107 117.64356 97.60396
#> Net_LnLx.Error Net_TnTx Net_TnTx.Error LxTx LxTx.Error Test_Dose
#> 1 143.03415 4697.495 69.24882 4.32801767 0.070695494 -1
#> 2 87.39498 4895.436 71.06916 1.53720681 0.028578369 -1
#> 3 123.18402 4858.079 71.66840 3.09871888 0.052275097 -1
#> 4 154.46828 4942.931 72.07168 4.77075371 0.076258410 -1
#> 5 176.39564 5569.158 75.88226 5.55525214 0.082052531 -1
#> 6 97.54912 5732.396 77.37976 1.61426805 0.027647946 -1
#> 7 14.86509 5989.356 78.87190 0.01629624 0.002491179 -1
#> UID
#> 1 5708d0a6ba7b4f22
#> 2 5708d0a6ba7b4f22
#> 3 5708d0a6ba7b4f22
#> 4 5708d0a6ba7b4f22
#> 5 5708d0a6ba7b4f22
#> 6 5708d0a6ba7b4f22
#> 7 5708d0a6ba7b4f22
## Run example with special case for
## the OTORX fit
if (FALSE) { # \dontrun{
results <- analyse_SAR.CWOSL(
object = object,
signal.integral.min = 1,
signal.integral.max = 2,
dose.points.test = 15,
background.integral.min = 900,
background.integral.max = 1000,
n.MC = 10,
fit.method = "OTORX")
} # }
##show De results
get_RLum(results)
#> De De.Error D01 D01.ERROR D02 D02.ERROR Dc D63 n_N De.MC
#> 1 1668.251 46.78683 1982.757 107.7542 NA NA NA NA 0.4854696 1664.089
#> Fit Mode HPDI68_L HPDI68_U HPDI95_L HPDI95_U .De.plot .De.raw
#> 1 EXP interpolation 1611.046 1708.69 1570.575 1763.294 1668.251 1668.251
#> RC.Status signal.range background.range signal.range.Tx background.range.Tx
#> 1 OK 1:2 900:1000 NA:NA NA:NA
#> ALQ POS GRAIN UID
#> 1 1 1 0 5708d0a6ba7b4f22
##show LnTnLxTx table
get_RLum(results, data.object = "LnLxTnTx.table")
#> Name Repeated Dose LnLx LnLx.BG TnTx TnTx.BG Net_LnLx
#> 1 Natural FALSE 0 20391 60.15842 4771 73.50495 20330.84158
#> 2 R1 FALSE 450 7591 65.70297 4977 81.56436 7525.29703
#> 3 R2 FALSE 1050 15150 96.17822 4960 101.92079 15053.82178
#> 4 R3 FALSE 2000 23700 118.49505 5064 121.06931 23581.50495
#> 5 R4 FALSE 2550 31094 155.92079 5715 145.84158 30938.07921
#> 6 R5 TRUE 450 9376 122.37624 5860 127.60396 9253.62376
#> 7 R0 FALSE 0 192 94.39604 6107 117.64356 97.60396
#> Net_LnLx.Error Net_TnTx Net_TnTx.Error LxTx LxTx.Error Test_Dose
#> 1 143.03415 4697.495 69.24882 4.32801767 0.070695494 -1
#> 2 87.39498 4895.436 71.06916 1.53720681 0.028578369 -1
#> 3 123.18402 4858.079 71.66840 3.09871888 0.052275097 -1
#> 4 154.46828 4942.931 72.07168 4.77075371 0.076258410 -1
#> 5 176.39564 5569.158 75.88226 5.55525214 0.082052531 -1
#> 6 97.54912 5732.396 77.37976 1.61426805 0.027647946 -1
#> 7 14.86509 5989.356 78.87190 0.01629624 0.002491179 -1
#> UID
#> 1 5708d0a6ba7b4f22
#> 2 5708d0a6ba7b4f22
#> 3 5708d0a6ba7b4f22
#> 4 5708d0a6ba7b4f22
#> 5 5708d0a6ba7b4f22
#> 6 5708d0a6ba7b4f22
#> 7 5708d0a6ba7b4f22
## Run example with special case for
## the OTORX fit
if (FALSE) { # \dontrun{
results <- analyse_SAR.CWOSL(
object = object,
signal.integral.min = 1,
signal.integral.max = 2,
dose.points.test = 15,
background.integral.min = 900,
background.integral.max = 1000,
n.MC = 10,
fit.method = "OTORX")
} # }